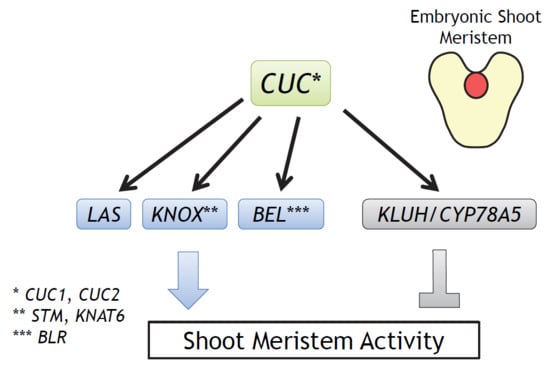
Establishment of the Embryonic Shoot Meristem Involves Activation of Two Classes of Genes with Opposing Functions for Meristem Activities
Abstract
:1. Introduction
2. Results
2.1. Selection of Candidate CUC1 and CUC2 Downstream Genes
2.2. Combined Expression of STM with LAS, BLR, and KNAT6 is Sufficient to Rescue the Embryonic Shoot Phenotypes of cuc1 cuc2
2.3. Combined Loss of Function of STM, LAS, BLR, and KNAT6 Severely Impairs Shoot Meristem Formation and Cotyledon Separation
2.4. The KLUH Gene Restricts the Embryonic Shoot Meristem and Counteracts STM
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Constructs
4.3. Rescue Experiments
4.4. Histological Analysis
4.5. DEX Induction and qRT-PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weigel, D.; Jürgens, G. Stem cells that make stems. Nature 2002, 415, 751–754. [Google Scholar] [CrossRef]
- Barton, M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010, 341, 95–113. [Google Scholar] [CrossRef] [Green Version]
- Gaillochet, C.; Lohmann, J.U. The never-ending story: From pluripotency to plant developmental plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [Green Version]
- Takada, S.; Hibara, K.; Ishida, T.; Tasaka, M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef] [Green Version]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of Phabulosa and Phavoluta in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [Green Version]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Tucker, E.; Hermann, M.; Laux, T. A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 2017, 40, 264–277.e264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knauer, S.; Holt, A.L.; Rubio-Somoza, I.; Tucker, E.J.; Hinze, A.; Pisch, M.; Javelle, M.; Timmermans, M.C.; Tucker, M.R.; Laux, T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 2013, 24, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aida, M.; Tasaka, M. Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 2006, 9, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Tasaka, M. Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol. Biol. 2006, 60, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Aida, M. Establishment of the embryonic shoot apical meristem in Arabidopsis thaliana. J. Plant Res. 2011, 124, 211–219. [Google Scholar] [CrossRef]
- Rast, M.I.; Simon, R. The meristem-to-organ boundary: More than an extremity of anything. Curr. Opin. Genet. Dev. 2008, 18, 287–294. [Google Scholar] [CrossRef]
- Wang, Q.; Hasson, A.; Rossmann, S.; Theres, K. Divide et impera: Boundaries shape the plant body and initiate new meristems. New Phytol. 2016, 209, 485–498. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Ishida, T.; Tasaka, M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 1999, 126, 1563–1570. [Google Scholar]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef]
- Belles-Boix, E.; Hamant, O.; Witiak, S.M.; Morin, H.; Traas, J.; Pautot, V. KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 2006, 18, 1900–1907. [Google Scholar] [CrossRef] [Green Version]
- Hibara, K.; Takada, S.; Tasaka, M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 2003, 36, 687–696. [Google Scholar] [CrossRef]
- Daimon, Y.; Takabe, K.; Tasaka, M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 2003, 44, 113–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant. J. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.H.; Zhang, X.N.; He, J.; Yu, H.P.; Wang, Y.; Shi, B.H.; Han, Y.Y.; Wang, G.X.; Feng, X.M.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Clarenz, O.; Schäfer, E.; Müller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastasiou, E.; Kenz, S.; Gerstung, M.; MacLean, D.; Timmer, J.; Fleck, C.; Lenhard, M. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 2007, 13, 843–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef] [Green Version]
- Aoyama, T.; Chua, N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997, 11, 605–612. [Google Scholar] [CrossRef]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Takano, S.; Niihama, M.; Smith, H.M.; Tasaka, M.; Aida, M. gorgon, a novel missense mutation in the SHOOT MERISTEMLESS gene, impairs shoot meristem homeostasis in Arabidopsis. Plant Cell Physiol. 2010, 51, 621–634. [Google Scholar] [CrossRef] [Green Version]
- Byrne, M.E.; Groover, A.T.; Fontana, J.R.; Martienssen, R.A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 2003, 130, 3941–3950. [Google Scholar] [CrossRef] [Green Version]
- Itoh, J.I.; Hasegawa, A.; Kitano, H.; Nagato, Y. A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell 1998, 10, 1511–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, K.; Ahn, B.O.; Kawakatsu, T.; Ito, Y.; Itoh, J.I.; Nagato, Y.; Kurata, N. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. USA 2004, 101, 875–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, U.; Grünewald, M.; Hobe, M.; Simon, R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002, 129, 565–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [Green Version]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [Green Version]
- Scofield, S.; Dewitte, W.; Nieuwland, J.; Murray, J.A.H. The Arabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 2013, 75, 53–66. [Google Scholar] [CrossRef]
- Kierzkowski, D.; Runions, A.; Vuolo, F.; Strauss, S.; Lymbouridou, R.; Routier-Kierzkowska, A.L.; Wilson-Sánchez, D.; Jenke, H.; Galinha, C.; Mosca, G.; et al. A growth-based framework for leaf shape development and diversity. Cell 2019, 177, 1405–1418.e1417. [Google Scholar] [CrossRef]
- Hake, S.; Smith, H.M.; Holtan, H.; Magnani, E.; Mele, G.; Ramirez, J. The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 2004, 20, 125–151. [Google Scholar] [CrossRef]
- Bhatt, A.M.; Etchells, J.P.; Canales, C.; Lagodienko, A.; Dickinson, H. VAAMANA—A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 2004, 328, 103–111. [Google Scholar] [CrossRef]
- Cole, M.; Nolte, C.; Werr, W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 2006, 34, 1281–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutjens, B.; Bao, D.; van Eck-Stouten, E.; Brand, M.; Smeekens, S.; Proveniers, M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009, 58, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wang, J.; Xiong, Y.; Yang, H.; Yang, M.; Ye, P.; Bencivenga, S.; Sablowski, R.; Jiao, Y. A self-activation loop maintains meristematic cell fate for branching. Curr. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tucker, D.J. Endogenous growth regulators in relation to side shoot development in the tomato. New Phytol. 1976, 77, 561–568. [Google Scholar] [CrossRef]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.Q.; Wang, J.G.; Wang, L.Y.; Wang, J.F.; Wang, Q.; Yu, P.; Bai, M.Y.; Fan, M. Gibberellin repression of axillary bud formation in Arabidopsis by modulation of DELLA-SPL9 complex activity. J. Integr. Plant Biol 2020, 62, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.M.; Hake, S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef] [Green Version]
- Sabatini, S.; Heidstra, R.; Wildwater, M.; Scheres, B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003, 17, 354–358. [Google Scholar] [CrossRef] [Green Version]
- Taoka, K.; Yanagimoto, Y.; Daimon, Y.; Hibara, K.; Aida, M.; Tasaka, M. The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J. 2004, 40, 462–473. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamski, N.M.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukaki, H.; Fujisawa, H.; Tasaka, M. SGR1, SGR2, SGR3: Novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant. Physiol. 1996, 110, 945–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Wolfe, D.S.; Nilsson, O.; Weigel, D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 1997, 7, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Sessions, A.; Weigel, D.; Yanofsky, M.F. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 1999, 20, 259–263. [Google Scholar] [CrossRef]






| Transgene | No Rescue a (%) | Mild Rescue b (%) | Strong Rescue c (%) | Total Number of T1 Seedlings |
|---|---|---|---|---|
| KLU | 100 | 0 | 0 | 25 |
| KNAT6 | 100 | 0 | 0 | 15 |
| UFO | 100 | 0 | 0 | 20 d |
| LAS | 86.5 | 13.5 | 0 | 37 |
| STM | 54.5 | 45.5 | 0 | 11 |
| PAN | 100 | 0 | 0 | 12 |
| LSH4 | 100 | 0 | 0 | 31 |
| BLR | 100 | 0 | 0 | 15 |
| SAI-LLP1 | 100 | 0 | 0 | 12 |
| ROXY20 | 100 | 0 | 0 | 12 |
| CUC2 | 8.3 | 50.0 | 41.7 | 12 |
| GUS | 100 | 0 | 0 | 4 |
| Transgene A | Transgene B | No Rescue a (%) | Mild Rescue b (%) | Strong Rescue c (%) | Total Number of F2 Seedlings | Group d |
|---|---|---|---|---|---|---|
| STM | - | 100 | 0 | 0 | 11 | a |
| - | LAS | 100 | 0 | 0 | 26 | a |
| STM | LAS | 0 | 71.1 | 28.9 | 38 | b |
| Transgene A | Transgene B | No Rescue a (%) | Mild Rescue b (%) | Strong Rescue c (%) | Total Number of F1 Seedlings |
|---|---|---|---|---|---|
| STM | − | 92.3 | 7.7 | 0 | 13 |
| − | KNAT6 | 100 | 0 | 0 | 6 |
| STM | KNAT6 | 42.9 | 57.1 | 0 | 14* |
| STM | − | 100 | 0 | 0 | 5 |
| - | UFO | 100 | 0 | 0 | 14 |
| STM | UFO | 100 | 0 | 0 | 12 |
| STM | − | 100 | 0 | 0 | 4 |
| − | PAN | 100 | 0 | 0 | 5 |
| STM | PAN | 100 | 0 | 0 | 9 |
| STM | − | 93.8 | 6.3 | 0 | 32 |
| − | LSH4 | 100 | 0 | 0 | 19 |
| STM | LSH4 | 92.9 | 7.1 | 0 | 14 |
| STM | − | 100 | 0 | 0 | 7 |
| − | BLR | 100 | 0 | 0 | 3 |
| STM | BLR | 16.7 | 0 | 83.3 | 6 ** |
| STM | − | 100 | 0 | 0 | 10 |
| − | GUS | 100 | 0 | 0 | 9 |
| STM | GUS | 100 | 0 | 0 | 10 |
| Genotype | Frequency of Fusion (%) | Total | Group * |
|---|---|---|---|
| Col | 0 | 55 | a |
| stm | 12.5 | 24 | b |
| blr stm | 91.3 | 23 | c |
| las stm | 35.3 | 17 | b |
| blr las stm | 95.7 | 23 | c |
| Genotype | Phenotype | Total Number of Seedlings | ||
|---|---|---|---|---|
| Normal a (%) | Weak b (%) | Strong c (%) | ||
| Col | 100 | 0 | 0 | 76 |
| klu | 100 * | 0 | 0 | 289 |
| cuc1 | 95.6 | 4.4 | 0 | 135 |
| cuc2 | 100 | 0 | 0 | 118 |
| klu cuc1 | 95.9 | 4.1 | 0 | 172 |
| klu cuc2 | 97.2 | 2.8 | 0 | 217 |
| cuc1 cuc2 | 0 | 0 | 100 | 50 |
| klu cuc1 cuc2 | 0 | 0 | 100 | 54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aida, M.; Tsubakimoto, Y.; Shimizu, S.; Ogisu, H.; Kamiya, M.; Iwamoto, R.; Takeda, S.; Karim, M.R.; Mizutani, M.; Lenhard, M.; et al. Establishment of the Embryonic Shoot Meristem Involves Activation of Two Classes of Genes with Opposing Functions for Meristem Activities. Int. J. Mol. Sci. 2020, 21, 5864. https://doi.org/10.3390/ijms21165864
Aida M, Tsubakimoto Y, Shimizu S, Ogisu H, Kamiya M, Iwamoto R, Takeda S, Karim MR, Mizutani M, Lenhard M, et al. Establishment of the Embryonic Shoot Meristem Involves Activation of Two Classes of Genes with Opposing Functions for Meristem Activities. International Journal of Molecular Sciences. 2020; 21(16):5864. https://doi.org/10.3390/ijms21165864
Chicago/Turabian StyleAida, Mitsuhiro, Yuka Tsubakimoto, Satoko Shimizu, Hiroyuki Ogisu, Masako Kamiya, Ryosuke Iwamoto, Seiji Takeda, Md Rezaul Karim, Masaharu Mizutani, Michael Lenhard, and et al. 2020. "Establishment of the Embryonic Shoot Meristem Involves Activation of Two Classes of Genes with Opposing Functions for Meristem Activities" International Journal of Molecular Sciences 21, no. 16: 5864. https://doi.org/10.3390/ijms21165864
APA StyleAida, M., Tsubakimoto, Y., Shimizu, S., Ogisu, H., Kamiya, M., Iwamoto, R., Takeda, S., Karim, M. R., Mizutani, M., Lenhard, M., & Tasaka, M. (2020). Establishment of the Embryonic Shoot Meristem Involves Activation of Two Classes of Genes with Opposing Functions for Meristem Activities. International Journal of Molecular Sciences, 21(16), 5864. https://doi.org/10.3390/ijms21165864