Double-Stranded RNA Binding Proteins in Serum Contribute to Systemic RNAi Across Phyla—Towards Finding the Missing Link in Achelata
Abstract
:1. Introduction
1.1. RNAi and Its Use in Decapods
1.2. Known Mechanisms of Systemic RNAi—A Drop in the Ocean
1.3. dsRNA Transport in Serum
2. Results
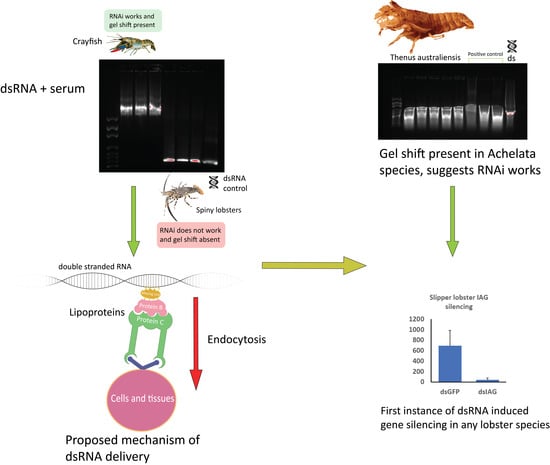
2.1. Correlation between Humoral Response to dsRNA and Silencing Capacity Using Exogenous dsRNA Across Phyla
2.2. Identification of Putative dsRNA Binding Protein Complex in Sera via Mass Spectrometry Analysis
2.3. Following the Gel Shift in Crustaceans
2.4. dsRNA-Induced IAG Knockdown in T. australiensis
3. Discussion
4. Materials and Methods
4.1. Hemolymph, Hemocoel, Plasma, and Serum Collection
4.2. Double-Stranded RNA Production
4.3. Electromobility Shift Assay (EMSA)
4.4. uHPLC Tandom QTof Mass Spectrometry Analysis
4.5. In Vivo T. Australiensis IAG Silencing
4.6. RNA Extraction and RT qPCR
4.7. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Rossi, J. RNAi mechanisms and applications. Biotechniques 2008, 44, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Sagi, A.; Manor, R.; Ventura, T. Gene silencing in crustaceans: From basic research to biotechnologies. Genes 2013, 4, 620–645. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Miesen, P.; Van Rij, R.P. Antiviral RNAi in Insects and Mammals: Parallels and Differences. Viruses 2019, 11, 448. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.-W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1191–1203. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Jiang, F.; Kalidas, S.; Smith, D.; Liu, Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 2006, 12, 1514–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakanishi, K. Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdisciplinary Reviews. RNA 2016, 7, 637–660. [Google Scholar]
- Liu, Y.; Ye, X.; Jiang, F.; Liang, C.; Chen, D.; Peng, J.; Kinch, L.N.; Grishin, N.V.; Liu, Q. C3PO, an Endoribonuclease That Promotes RNAi by Facilitating RISC Activation. Science 2009, 325, 750–753. [Google Scholar] [CrossRef] [Green Version]
- Ming, D.; Wall, M.E.; Sanbonmatsu, K.Y. Domain motions of Argonaute, the catalytic engine of RNA interference. BMC Bioinform. 2007, 8, 470. [Google Scholar] [CrossRef] [Green Version]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A Sexual Shift Induced by Silencing of a Single Insulin-Like Gene in Crayfish: Ovarian Upregulation and Testicular Degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef] [Green Version]
- Pamuru, R.R.; Rosen, O.; Manor, R.; Chung, J.S.; Zmora, N.; Glazer, L.; Aflalo, E.D.; Weil, S.; Tamone, S.L.; Sagi, A. Stimulation of molt by RNA interference of the molt-inhibiting hormone in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 2012, 178, 227–236. [Google Scholar] [CrossRef]
- De Santis, C.; Wade, N.M.; Jerry, D.R.; Preston, N.P.; Glencross, B.D.; Sellars, M.J. Growing backwards: An inverted role for the shrimp ortholog of vertebrate myostatin and GDF11. J. Exp. Biol. 2011, 214, 2671–2677. [Google Scholar] [CrossRef] [Green Version]
- Ponprateep, S.; Tharntada, S.; Somboonwiwat, K.; Tassanakajon, A. Gene silencing reveals a crucial role for anti-lipopolysaccharide factors from Penaeus monodon in the protection against microbial infections. Fish Shellfish Immunol. 2012, 32, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Ruvkun, G. New insights into siRNA amplification and RNAi. RNA Biol. 2012, 9, 1045–1049. [Google Scholar] [CrossRef] [Green Version]
- Feinberg, E.H.; Hunter, C.P. Transport of dsRNA into Cells by the Transmembrane Protein SID-1. Science 2003, 301, 1545–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynant, N.; Santos, D.; Van Wielendaele, P.; Vanden Broeck, J. Scavenger receptor-mediated endocytosis facilitates RNA interference in the desert locust, Schistocerca gregaria. Insect Mol. Biol. 2014, 23, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Han, Z. Cloning and Phylogenetic Analysis of Sid-1-Like Genes from Aphids. J. Insect Sci. 2008, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.-H.; He, J.-G. Chapter Six—Nucleic acid sensing in invertebrate antiviral immunity. In International Review of Cell and Molecular Biology; Vanpouille-Box, C., Galluzzi, L., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 345, pp. 287–360. [Google Scholar]
- Maruekawong, K.; Tirasophon, W.; Panyim, S.; Attasart, P. Involvement of Lv SID-1 in dsRNA uptake in Litopenaeus vannamei. Aquaculture 2018, 482, 65–72. [Google Scholar] [CrossRef]
- Shpak, N.; Manor, R.; Abilevich, L.K.; Mantal, O.; Shavit, K.; Aflalo, E.D.; Toiber, D.; Sagi, A. Short versus long double-stranded RNA activation of a post-transcriptional gene knockdown pathway. RNA Biol. 2017, 14, 1766–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McEwan, D.L.; Weisman, A.S.; Hunter, C.P. Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell 2012, 47, 746–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winston, W.M.; Sutherlin, M.; Wright, A.J.; Feinberg, E.H.; Hunter, C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 2007, 104, 10565–10570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinheiro, D.H.; Vélez, A.M.; Fishilevich, E.; Wang, H.; Carneiro, N.P.; Valencia-Jiménez, A.; Valicente, F.H.; Narva, K.E.; Siegfried, B.D. Clathrin-dependent endocytosis is associated with RNAi response in the western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS ONE 2018, 13, e0201849. [Google Scholar] [CrossRef] [Green Version]
- Tatematsu, M.; Funami, K.; Seya, T.; Matsumoto, M. Extracellular RNA Sensing by Pattern Recognition Receptors. J. Innate Immun. 2018, 10, 398–406. [Google Scholar] [CrossRef]
- Xiao, D.; Gao, X.; Xu, J.; Liang, X.; Li, Q.; Yao, J.; Zhu, K.Y. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle. Insect Biochem. Mol. Biol. 2015, 60, 68–77. [Google Scholar] [CrossRef]
- Saleh, M.-C.; van Rij, R.P.; Hekele, A.; Gillis, A.; Foley, E.; O’Farrell, P.H.; Andino, R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 2006, 8, 793–802. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, X.; Yu, D.; Kang, L. The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA Biol. 2012, 9, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.; Gatehouse, J.; Fitches, E. A Systematic Study of RNAi Effects and dsRNA Stability in Tribolium castaneum and Acyrthosiphon pisum, Following Injection and Ingestion of Analogous dsRNAs. Int. J. Mol. Sci. 2018, 19, 1079. [Google Scholar] [CrossRef] [Green Version]
- Maruekawong, K.; Panyim, S.; Attasart, P. Involvement of endocytosis in cellular uptake of injected dsRNA into hepatopancreas but not in gill of Litopenaeus vannamei. Aquaculture 2019, 500, 393–397. [Google Scholar] [CrossRef]
- Attasart, P.; Namramoon, O.; Kongphom, U.; Chimwai, C.; Panyim, S. Ingestion of bacteria expressing dsRNA triggers specific RNA silencing in shrimp. Virus Res. 2013, 171, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Sakashita, K.; Tatsuke, T.; Masaki, Y.; Jae-Man, L.; Kawaguchi, Y.; Kusakabe, T. dsRNA Binding Activity of Silworm Larval Hemolymph is Mediated by Lipophorin Complex. J. Fac. Agric. Kyushu Univ. 2009, 54, 401–406. [Google Scholar]
- McMahon, K.M.; Thaxton, C.S. High-density lipoproteins for the systemic delivery of short interfering RNA. Expert Opin. Drug Deliv. 2014, 11, 231–247. [Google Scholar] [CrossRef] [Green Version]
- Wynant, N.; Duressa, T.F.; Santos, D.; Van Duppen, J.; Proost, P.; Huybrechts, R.; Vanden Broeck, J. Lipophorins can adhere to dsRNA, bacteria and fungi present in the hemolymph of the desert locust: A role as general scavenger for pathogens in the open body cavity. J. Insect Physiol. 2014, 64, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Maori, E.; Garbian, Y.; Kunik, V.; Mozes-Koch, R.; Malka, O.; Kalev, H.; Sabath, N.; Sela, I.; Shafir, S. A Transmissible RNA Pathway in Honey Bees. Cell Rep. 2019, 27, 1949–1959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maori, E.; Navarro, I.C.; Boncristiani, H.; Seilly, D.J.; Rudolph, K.L.M.; Sapetschnig, A.; Lin, C.-C.; Ladbury, J.E.; Evans, J.D.; Heeney, J.L.; et al. A Secreted RNA Binding Protein Forms RNA-Stabilizing Granules in the Honeybee Royal Jelly. Mol. Cell 2019, 74, 598–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mingels, L.; Wynant, N.; Santos, D.; Peeters, P.; Gansemans, Y.; Billen, J.; Van Nieuwerburgh, F.; Broeck, J.V. Extracellular vesicles spread the RNA interference signal of Tribolium castaneum TcA cells. Insect Biochem. Mol. Biol. 2020, 122, 103377. [Google Scholar] [CrossRef] [PubMed]
- Tassetto, M.; Kunitomi, M.; Andino, R. Circulating Immune Cells Mediate a Systemic RNAi-Based Adaptive Antiviral Response in Drosophila. Cell 2017, 169, 314–325. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing Sexual Differentiation: Full Functional Sex Reversal Achieved through Silencing of a Single Insulin-Like Gene in the Prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 90–91. [Google Scholar] [CrossRef]
- Su, F.J.; Lu, M.-W. Application and efficacy test of siRNA vaccine against nervous necrosis virus infection. Fish Shellfish Immunol. 2016, 53, 97. [Google Scholar] [CrossRef]
- Setten, R.L.; Rossi, J.J.; Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Caplen, N.J.; Parrish, S.; Imani, F.; Fire, A.; Morgan, R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 2001, 98, 9742–9747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, D.; Li, Q.; Yu, H. RNA Interference by Ingested dsRNA-Expressing Bacteria to Study Shell Biosynthesis and Pigmentation in Crassostrea gigas. Mar. Biotechnol. 2019, 21, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zeng, B.; Ling, L.; Xu, J.; You, L.; Aslam, A.F.M.; Tan, A.; Huang, Y. Enhancement of Larval RNAi Efficiency by Over-expressing Argonaute2 in Bombyx mori. Int. J. Biol. Sci. 2015, 11, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Gleghorn, M.L.; Maquat, L.E. ‘Black sheep’ that don’t leave the double-stranded RNA-binding domain fold. Trends Biochem. Sci. 2014, 39, 328–340. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-T.; Chang, C.-I.; Hseu, J.-R.; Liu, K.-F.; Tsai, J.-M. Immune responses of prophenoloxidase and cytosolic manganese superoxide dismutase in the freshwater crayfish Cherax quadricarinatus against a virus and bacterium. Mol. Immunol. 2013, 56, 72–80. [Google Scholar] [CrossRef]
- Jorieux, S.; Fressinaud, E.; Goudemand, J.; Gaucher, C.; Meyer, D.; Mazurier, C. Conformational changes in the D’ domain of von Willebrand factor induced by CYS 25 and CYS 95 mutations lead to factor VIII binding defect and multimeric impairment. Blood 2000, 95, 3139–3145. [Google Scholar] [CrossRef]
- Vazquez, L.; Alpuche, J.; Maldonado, G.; Agundis, C.; Pereyra-Morales, A.; Zenteno, E. Review: Immunity mechanisms in crustaceans. Innate Immun. 2009, 15, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.P.; Behringer, D.C.; Bojko, J. White Spot Syndrome Virus and the Caribbean Spiny Lobster, Panulirus argus: Susceptibility and Behavioral Immunity. J. Invertebr. Pathol. 2019, 162, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shields, J.; Behringer, D. A new pathogenic virus in the Caribbean spiny lobster Panulirus argus from the Florida Keys. Diseases of Aquatic Organisms 2004, 59, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Hyde, C.J.; Fitzgibbon, Q.P.; Elizur, A.; Smith, G.G.; Ventura, T. Transcriptional profiling of spiny lobster metamorphosis reveals three new additions to the nuclear receptor superfamily. BMC Genom. 2019, 20, 531. [Google Scholar]
- Song, M.-S.; Rossi, J.J. Molecular mechanisms of Dicer: Endonuclease and enzymatic activity. Biochem. J. 2017, 474, 1603–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagnon, K.T.; Corey, D.R. Argonaute and the Nuclear RNAs: New Pathways for RNA-Mediated Control of Gene Expression. Nucleic Acid Ther. 2012, 22, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.F.; Puppione, D.L. Serum lipoproteins in the spiny lobster, Panulirus interruptus. Comp. Biochem. Physiol. Part B Comp. Biochem. 1978, 59, 239–243. [Google Scholar] [CrossRef]
- Kopacek, P.; Hall, M.; Soderhall, K. Characterization of a clotting protein, isolated from plasma of the freshwater crayfish Pacifastacus leniusculus. Eur. J. Biochem. 1993, 213, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wyeth, R.C.; Liang, D.; Bose, U.; Ni, G.; McManus, D.P.; Cummins, S.F. A Biomphalaria glabrata peptide that stimulates significant behaviour modifications in aquatic free-living Schistosoma mansoni miracidia. PLoS Negl. Trop. Dis. 2019, 13, e0006948. [Google Scholar] [CrossRef]
- Chieu, H.D.; Turner, L.; Smith, M.K.; Wang, T.; Nocillado, J.; Palma, P.; Suwansa-Ard, S.; Elizur, A.; Cummins, S.F. Aquaculture Breeding Enhancement: Maturation and Spawning in Sea Cucumbers Using a Recombinant Relaxin-Like Gonad-Stimulating Peptide. Front. Genet. 2019, 10, 77. [Google Scholar] [CrossRef] [Green Version]
- Whitten, M.M.A.; Tew, I.F.; Lee, B.L.; Ratcliffe, N.A. A Novel Role for an Insect Apolipoprotein (Apolipophorin III) in β-1,3-Glucan Pattern Recognition and Cellular Encapsulation Reactions. J. Immunol. 2004, 172, 2177–2185. [Google Scholar] [CrossRef] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Cummins, S.F.; Elizur, A.; Ventura, T. Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep. 2016, 6, 38658. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Stewart, M.J.; Chandler, J.C.; Rotgans, B.; Elizur, A.; Hewitt, A.W. Molecular aspects of eye development and regeneration in the Australian redclaw crayfish, Cherax quadricarinatus. Aquac. Fish. 2018, 4, 27–36. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, L.; Zhang, F.; Huang, S.; Merchant, A.; Zhou, X.; Li, Z. Over-expression of RNA interference (RNAi) core machinery improves susceptibility to RNAi in silkworm larvae. Insect Mol. Biol. 2020, 29, 353–362. [Google Scholar] [CrossRef]
- Yang, C.; Wu, F.; Lu, X.; Jiang, M.; Liu, W.; Yu, L.; Tian, J.; Wen, H. Growth arrest specific gene 2 in tilapia (Oreochromis niloticus): Molecular characterization and functional analysis under low-temperature stress. BMC Mol. Biol. 2017, 18, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.; An, X.; Jiang, Y.-D.; Ding, B.-Y.; Shang, F.; Christiaens, O.; Taning, C.N.T.; Smagghe, G.; Niu, J.; Wang, J.-J. Induction of RNAi Core Machinery’s Gene Expression by Exogenous dsRNA and the Effects of Pre-exposure to dsRNA on the Gene Silencing Efficiency in the Pea Aphid (Acyrthosiphon pisum). Front. Physiol. 2019, 9, 1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wu, X.; Chen, T.; Jiang, X.; Ren, C. Molecular characterization, inducible expression and functional analysis of an IKKβ from the tropical sea cucumber Holothuria leucospilota. Fish Shellfish Immunol. 2020, 104, 622–632. [Google Scholar] [CrossRef]
- Srinivas, R.V.; Birkedal, B.; Owens, R.J.; Anantharamaiah, G.M.; Segrest, J.P.; Compans, R.W. Antiviral effects of apolipoprotein A-I and its synthetic amphipathic peptide analogs. Virology 1990, 176, 48–57. [Google Scholar] [CrossRef]
- Fukunaga, R.; Doudna, J.A. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009, 28, 545–555. [Google Scholar] [CrossRef] [Green Version]








| Species | Proteins Identified (via BlastP) | No. of Peptides | Sequence Coverage | e-Value | % Identity |
|---|---|---|---|---|---|
| C. quadricarinatus | clotting protein precursor [Pacifastacus leniusculus] | 1 | 1 | 0 | 50% |
| E. lanceolatus | apolipoprotein A-Ib [Epinephelus lanceolatus] | 10 | 24 | 0 | 99.62% |
| E. lanceolatus | 14 kDa apolipoprotein, partial [Epinephelus bruneus] | 7 | 47 | 1 × 10−87 | 89.51 |
| B. mori | apolipophorin III precursor [Bombyx mori] | 2 | 14 | 7 × 10−131 | 100% |
| B. mori | low molecular 30 kDa lipoprotein PBMHP-6 [Bombyx mandarina] | 1 | 4 | 0 | 97.66% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banks, T.M.; Wang, T.; Fitzgibbon, Q.P.; Smith, G.G.; Ventura, T. Double-Stranded RNA Binding Proteins in Serum Contribute to Systemic RNAi Across Phyla—Towards Finding the Missing Link in Achelata. Int. J. Mol. Sci. 2020, 21, 6967. https://doi.org/10.3390/ijms21186967
Banks TM, Wang T, Fitzgibbon QP, Smith GG, Ventura T. Double-Stranded RNA Binding Proteins in Serum Contribute to Systemic RNAi Across Phyla—Towards Finding the Missing Link in Achelata. International Journal of Molecular Sciences. 2020; 21(18):6967. https://doi.org/10.3390/ijms21186967
Chicago/Turabian StyleBanks, Thomas M., Tianfang Wang, Quinn P. Fitzgibbon, Gregory G. Smith, and Tomer Ventura. 2020. "Double-Stranded RNA Binding Proteins in Serum Contribute to Systemic RNAi Across Phyla—Towards Finding the Missing Link in Achelata" International Journal of Molecular Sciences 21, no. 18: 6967. https://doi.org/10.3390/ijms21186967
APA StyleBanks, T. M., Wang, T., Fitzgibbon, Q. P., Smith, G. G., & Ventura, T. (2020). Double-Stranded RNA Binding Proteins in Serum Contribute to Systemic RNAi Across Phyla—Towards Finding the Missing Link in Achelata. International Journal of Molecular Sciences, 21(18), 6967. https://doi.org/10.3390/ijms21186967