Nicotinic Receptor Subunits Atlas in the Adult Human Lung
Abstract
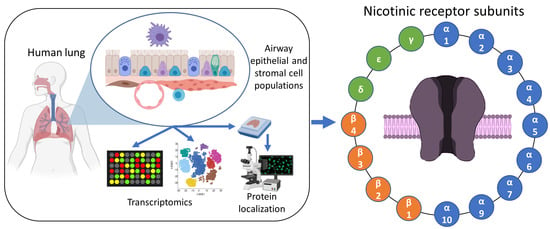
:1. Introduction
2. Results
2.1. Smoking-Associated Pulmonary nAChR Subunit Transcript Expressions
2.2. Differential Pulmonary nAChRs Transcript Expressions at the Single-Cell Scale
2.3. Identification of nAChR Subunits in Bronchial and Large Bronchiolar Epithelia
3. Discussion
4. Materials and Methods
4.1. Human Subjects
4.2. Sample Processing
4.3. RT-qPCR Analyses
4.4. Immunofluorescent Staining and Analyses
4.5. Transcriptome Profiling Microarray Analysis
4.6. Single-Cell Sequencing
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| nAChRs | Nicotinic acetylcholine receptors |
| AEC | Airway epithelial cells |
| COPD | Chronic obstructive pulmonary disease |
| COVID-19 | Coronavirus disease 2019 |
| ACE-2 | Angiotensin-converting enzyme-2 |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus |
| FFPE | Formalin-fixed paraffin-embedded |
| LAEC | Large airway epithelial cell |
| SAEC | Small airway epithelial cell |
| SNP | Single-nucleotide polymorphisms |
| RAS | Renin–angiotensin system |
References
- Zoli, M.; Pucci, S.; Vilella, A.; Gotti, C. Neuronal and Extraneuronal Nicotinic Acetylcholine Receptors. Curr. Neuropharmacol. 2018, 16, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Dani, J.A. Neuronal Nicotinic Acetylcholine Receptor Structure and Function and Response to Nicotine. Int. Rev. Neurobiol. 2015, 124, 3–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gündisch, D.; Eibl, C. Nicotinic acetylcholine receptor ligands, a patent review (2006–2011). Expert Opin. Ther. Pat. 2011, 21, 1867–1896. [Google Scholar] [CrossRef] [Green Version]
- Grando, S.A. Connections of nicotine to cancer. Nat. Rev. Cancer 2014, 14, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Grassi, F.; Fucile, S. Calcium influx through muscle nAChR-channels: One route, multiple roles. Neuroscience 2019, 439, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Conti-Tronconi, B.M.; McLane, K.E.; Raftery, M.A.; Grando, S.A.; Protti, M.P. The nicotinic acetylcholine receptor: Structure and autoimmune pathology. Crit. Rev. Biochem. Mol. Biol. 1994, 29, 69–123. [Google Scholar] [CrossRef]
- Rudolf, R.; Straka, T. Nicotinic acetylcholine receptor at vertebrate motor endplates: Endocytosis, recycling, and degradation. Neurosci. Lett. 2019, 711, 134434. [Google Scholar] [CrossRef]
- Kabbani, N.; Nichols, R.A. Beyond the Channel: Metabotropic Signaling by Nicotinic Receptors. Trends Pharmacol. Sci. 2018, 39, 354–366. [Google Scholar] [CrossRef]
- Papke, R.L.; Lindstrom, J.M. Nicotinic acetylcholine receptors: Conventional and unconventional ligands and signaling. Neuropharmacology 2020, 168, 108021. [Google Scholar] [CrossRef]
- Albuquerque, E.X.; Pereira, E.F.R.; Alkondon, M.; Rogers, S.W. Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 2009, 89, 73–120. [Google Scholar] [CrossRef] [Green Version]
- Baranowska, U.; Wiśniewska, R.J. The α7-nACh nicotinic receptor and its role in memory and selected diseases of the central nervous system. Postepy. Hig. Med. Dosw. (Online) 2017, 71, 633–648. [Google Scholar] [CrossRef]
- Bertrand, D.; Terry, A.V. The wonderland of neuronal nicotinic acetylcholine receptors. Biochem. Pharmacol. 2018, 151, 214–225. [Google Scholar] [CrossRef]
- Crespi, A.; Colombo, S.F.; Gotti, C. Proteins and chemical chaperones involved in neuronal nicotinic receptor expression and function: An update. Br. J. Pharmacol. 2018, 175, 1869–1879. [Google Scholar] [CrossRef]
- Medjber, K.; Freidja, M.L.; Grelet, S.; Lorenzato, M.; Maouche, K.; Nawrocki-Raby, B.; Birembaut, P.; Polette, M.; Tournier, J.-M. Role of nicotinic acetylcholine receptors in cell proliferation and tumour invasion in broncho-pulmonary carcinomas. Lung Cancer 2015, 87, 258–264. [Google Scholar] [CrossRef]
- Koukouli, F.; Rooy, M.; Changeux, J.-P.; Maskos, U. Nicotinic receptors in mouse prefrontal cortex modulate ultraslow fluctuations related to conscious processing. Proc. Natl. Acad. Sci. USA 2016, 113, 14823–14828. [Google Scholar] [CrossRef] [Green Version]
- Changeux, J.-P.; Corringer, P.-J.; Maskos, U. The nicotinic acetylcholine receptor: From molecular biology to cognition. Neuropharmacology 2015, 96, 135–136. [Google Scholar] [CrossRef]
- Le Novère, N.; Corringer, P.-J.; Changeux, J.-P. The diversity of subunit composition in nAChRs: Evolutionary origins, physiologic and pharmacologic consequences. J. Neurobiol. 2002, 53, 447–456. [Google Scholar] [CrossRef]
- Wu, J.; Liu, Q.; Tang, P.; Mikkelsen, J.D.; Shen, J.; Whiteaker, P.; Yakel, J.L. Heteromeric α7β2 Nicotinic Acetylcholine Receptors in the Brain. Trends Pharmacol. Sci. 2016, 37, 562–574. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Cheuk, I.W.Y.; Shin, V.Y.; Kwong, A. Acetylcholine receptors: Key players in cancer development. Surg. Oncol. 2019, 31, 46–53. [Google Scholar] [CrossRef]
- Maouche, K.; Polette, M.; Jolly, T.; Medjber, K.; Cloëz-Tayarani, I.; Changeux, J.-P.; Burlet, H.; Terryn, C.; Coraux, C.; Zahm, J.-M.; et al. {alpha}7 nicotinic acetylcholine receptor regulates airway epithelium differentiation by controlling basal cell proliferation. Am. J. Pathol. 2009, 175, 1868–1882. [Google Scholar] [CrossRef] [Green Version]
- Munakata, S.; Ishimori, K.; Kitamura, N.; Ishikawa, S.; Takanami, Y.; Ito, S. Oxidative stress responses in human bronchial epithelial cells exposed to cigarette smoke and vapor from tobacco- and nicotine-containing products. Regul. Toxicol. Pharmacol. 2018, 99, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.; Tarran, R. E-cigarettes, nicotine, the lung and the brain: Multi-level cascading pathophysiology. J. Physiol. 2020, JP278388. [Google Scholar] [CrossRef]
- Kabbani, N.; Nordman, J.C.; Corgiat, B.A.; Veltri, D.P.; Shehu, A.; Seymour, V.A.; Adams, D.J. Are nicotinic acetylcholine receptors coupled to G proteins? Bioessays 2013, 35, 1025–1034. [Google Scholar] [CrossRef]
- King, J.R.; Ullah, A.; Bak, E.; Jafri, M.S.; Kabbani, N. Ionotropic and Metabotropic Mechanisms of Allosteric Modulation of α7 Nicotinic Receptor Intracellular Calcium. Mol. Pharmacol. 2018, 93, 601–611. [Google Scholar] [CrossRef] [Green Version]
- Cui, K.; Ge, X.; Ma, H. Four SNPs in the CHRNA3/5 alpha-neuronal nicotinic acetylcholine receptor subunit locus are associated with COPD risk based on meta-analyses. PLoS ONE 2014, 9, e102324. [Google Scholar] [CrossRef]
- Maskos, U. The nicotinic receptor alpha5 coding polymorphism rs16969968 as a major target in disease: Functional dissection and remaining challenges. J. Neurochem. 2020. [Google Scholar] [CrossRef] [Green Version]
- Sriram, K.; Insel, P.A. A hypothesis for pathobiology and treatment of COVID-19: The centrality of ACE1/ACE2 imbalance. Br. J. Pharmacol. 2020, bph.15082. [Google Scholar] [CrossRef]
- Oakes, J.M.; Fuchs, R.M.; Gardner, J.D.; Lazartigues, E.; Yue, X. Nicotine and the renin-angiotensin system. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 315, R895–R906. [Google Scholar] [CrossRef] [Green Version]
- Changeux, J.-P.; Amoura, Z.; Rey, F.A.; Miyara, M. A nicotinic hypothesis for Covid-19 with preventive and therapeutic implications. Comptes Rendus Biol. 2020, 343, 33–39. [Google Scholar] [CrossRef]
- Lam, D.C.-L.; Luo, S.Y.; Fu, K.-H.; Lui, M.M.-S.; Chan, K.-H.; Wistuba, I.I.; Gao, B.; Tsao, S.-W.; Ip, M.S.-M.; Minna, J.D. Nicotinic acetylcholine receptor expression in human airway correlates with lung function. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 310, L232–L239. [Google Scholar] [CrossRef] [Green Version]
- Reyfman, P.A.; Walter, J.M.; Joshi, N.; Anekalla, K.R.; McQuattie-Pimentel, A.C.; Chiu, S.; Fernandez, R.; Akbarpour, M.; Chen, C.-I.; Ren, Z.; et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1517–1536. [Google Scholar] [CrossRef] [PubMed]
- Schiller, H.B.; Montoro, D.T.; Simon, L.M.; Rawlins, E.L.; Meyer, K.B.; Strunz, M.; Vieira Braga, F.A.; Timens, W.; Koppelman, G.H.; Budinger, G.R.S.; et al. The Human Lung Cell Atlas: A High-Resolution Reference Map of the Human Lung in Health and Disease. Am. J. Respir. Cell Mol. Biol. 2019, 61, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Vieira Braga, F.A.; Kar, G.; Berg, M.; Carpaij, O.A.; Polanski, K.; Simon, L.M.; Brouwer, S.; Gomes, T.; Hesse, L.; Jiang, J.; et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019, 25, 1153–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaragosi, L.E.; Deprez, M.; Barbry, P. Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract. Biochem. Soc. Trans. 2020, 48, 327–336. [Google Scholar] [CrossRef]
- Ruiz García, S.; Deprez, M.; Lebrigand, K.; Cavard, A.; Paquet, A.; Arguel, M.-J.; Magnone, V.; Truchi, M.; Caballero, I.; Leroy, S.; et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 2019. [Google Scholar] [CrossRef] [Green Version]
- Moser, N.; Mechawar, N.; Jones, I.; Gochberg-Sarver, A.; Orr-Urtreger, A.; Plomann, M.; Salas, R.; Molles, B.; Marubio, L.; Roth, U.; et al. Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures: Evaluation of nicotinic receptor antibodies. J. Neurochem. 2007, 102, 479–492. [Google Scholar] [CrossRef]
- Rommel, F.R.; Raghavan, B.; Paddenberg, R.; Kummer, W.; Tumala, S.; Lochnit, G.; Gieler, U.; Peters, E.M.J. Suitability of Nicotinic Acetylcholine Receptor α7 and Muscarinic Acetylcholine Receptor 3 Antibodies for Immune Detection: Evaluation in Murine Skin. J. Histochem. Cytochem. 2015, 63, 329–339. [Google Scholar] [CrossRef]
- Lam, D.C.-L.; Girard, L.; Ramirez, R.; Chau, W.-S.; Suen, W.-S.; Sheridan, S.; Tin, V.P.C.; Chung, L.-P.; Wong, M.P.; Shay, J.W.; et al. Expression of Nicotinic Acetylcholine Receptor Subunit Genes in Non-Small-Cell Lung Cancer Reveals Differences between Smokers and Nonsmokers. Cancer Res. 2007, 67, 4638–4647. [Google Scholar] [CrossRef] [Green Version]
- Garg, B.K.; Loring, R.H. Evaluating Commercially Available Antibodies for Rat α7 Nicotinic Acetylcholine Receptors. J. Histochem. Cytochem. 2017, 65, 499–512. [Google Scholar] [CrossRef] [Green Version]
- Barrantes, F.J. Cell-surface translational dynamics of nicotinic acetylcholine receptors. Front. Synaptic Neurosci. 2014, 6. [Google Scholar] [CrossRef] [Green Version]
- Amos, C.I.; Wu, X.; Broderick, P.; Gorlov, I.P.; Gu, J.; Eisen, T.; Dong, Q.; Zhang, Q.; Gu, X.; Vijayakrishnan, J.; et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008, 40, 616–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, R.J.; McKay, J.D.; Gaborieau, V.; Boffetta, P.; Hashibe, M.; Zaridze, D.; Mukeria, A.; Szeszenia-Dabrowska, N.; Lissowska, J.; Rudnai, P.; et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008, 452, 633–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Li, L.; Zhao, C.; Pan, M.; Qian, Z.; Su, X. Deficiency of α7 Nicotinic Acetylcholine Receptor Attenuates Bleomycin-Induced Lung Fibrosis in Mice. Mol. Med. 2017, 23, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-J.; Jia, Y.-F.; Ma, X.-L. Alpha5 Nicotinic Acetylcholine Receptor Contributes to Nicotine-Induced Lung Cancer Development and Progression. Front. Pharmacol. 2017, 8, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Ma, X. α5-nAChR modulates nicotine-induced cell migration and invasion in A549 lung cancer cells. Exp. Toxicol. Pathol. 2015, 67, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Bordas, A.; Cedillo, J.L.; Arnalich, F.; Esteban-Rodriguez, I.; Guerra-Pastrián, L.; de Castro, J.; Martín-Sánchez, C.; Atienza, G.; Fernández-Capitan, C.; Rios, J.J.; et al. Expression patterns for nicotinic acetylcholine receptor subunit genes in smoking-related lung cancers. Oncotarget 2017, 8, 67878–67890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, J.; Liu, Y.; Sun, Z.; Zhangsun, D.; Luo, S. Identification of nicotinic acetylcholine receptor subunits in different lung cancer cell lines and the inhibitory effect of alpha-conotoxin TxID on lung cancer cell growth. Eur. J. Pharmacol. 2019, 865, 172674. [Google Scholar] [CrossRef]
- Witayateeraporn, W.; Arunrungvichian, K.; Pothongsrisit, S.; Doungchawee, J.; Vajragupta, O.; Pongrakhananon, V. α7-Nicotinic acetylcholine receptor antagonist QND7 suppresses non-small cell lung cancer cell proliferation and migration via inhibition of Akt/mTOR signaling. Biochem. Biophys. Res. Commun. 2020, 521, 977–983. [Google Scholar] [CrossRef]
- Friedman, J.R.; Richbart, S.D.; Merritt, J.C.; Brown, K.C.; Nolan, N.A.; Akers, A.T.; Lau, J.K.; Robateau, Z.R.; Miles, S.L.; Dasgupta, P. Acetylcholine signaling system in progression of lung cancers. Pharmacol. Ther. 2019, 194, 222–254. [Google Scholar] [CrossRef]
- Pillai, S.G.; Ge, D.; Zhu, G.; Kong, X.; Shianna, K.V.; Need, A.C.; Feng, S.; Hersh, C.P.; Bakke, P.; Gulsvik, A.; et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): Identification of two major susceptibility loci. PLoS Genet. 2009, 5, e1000421. [Google Scholar] [CrossRef] [Green Version]
- Pillai, S.G.; Kong, X.; Edwards, L.D.; Cho, M.H.; Anderson, W.H.; Coxson, H.O.; Lomas, D.A.; Silverman, E.K. Loci Identified by Genome-wide Association Studies Influence Different Disease-related Phenotypes in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2010, 182, 1498–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, M.H.; McDonald, M.-L.N.; Zhou, X.; Mattheisen, M.; Castaldi, P.J.; Hersh, C.P.; DeMeo, D.L.; Sylvia, J.S.; Ziniti, J.; Laird, N.M.; et al. Risk loci for chronic obstructive pulmonary disease: A genome-wide association study and meta-analysis. Lancet Respir. Med. 2014, 2, 214–225. [Google Scholar] [CrossRef] [Green Version]
- The Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010, 42, 441–447. [CrossRef] [PubMed] [Green Version]
- Douaoui, S.; Djidjik, R.; Boubakeur, M.; Ghernaout, M.; Touil-boukoffa, C.; Oumouna, M.; Derrar, F.; Amrani, Y. GTS-21, an α7nAChR agonist, suppressed the production of key inflammatory mediators by PBMCs that are elevated in COPD patients and associated with impaired lung function. Immunobiology 2020, 225, 151950. [Google Scholar] [CrossRef]
- Bray, M.J.; Chen, L.; Fox, L.; Hancock, D.B.; Culverhouse, R.C.; Hartz, S.M.; Johnson, E.O.; Liu, M.; McKay, J.D.; Saccone, N.L.; et al. Dissecting the genetic overlap of smoking behaviors, lung cancer, and chronic obstructive pulmonary disease: A focus on nicotinic receptors and nicotine metabolizing enzyme. Genet. Epidemiol. 2020, 44, 748–758. [Google Scholar] [CrossRef]
- Yamada, M.; Ichinose, M. The Cholinergic Pathways in Inflammation: A Potential Pharmacotherapeutic Target for COPD. Front. Pharmacol. 2018, 9, 1426. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Bossé, Y.; Xiao, F.; Kheradmand, F.; Amos, C.I. Tobacco Smoking Increases the Lung Gene Expression of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, rccm.202003-0693LE. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Rostami, M.R.; Leopold, P.L.; Mezey, J.G.; O’Beirne, S.L.; Strulovici-Barel, Y.; Crystal, R.G. Expression of the SARS-CoV-2 ACE2 Receptor in the Human Airway Epithelium. Am. J. Respir. Crit. Care Med. 2020, rccm.202003-0541OC. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Russo, P.; Bonassi, S.; Giacconi, R.; Malavolta, M.; Tomino, C.; Maggi, F. COVID-19 and smoking: Is nicotine the hidden link? Eur. Respir. J. 2020, 55, 2001116. [Google Scholar] [CrossRef]
- Du Rand, I.A.; Blaikley, J.; Booton, R.; Chaudhuri, N.; Gupta, V.; Khalid, S.; Mandal, S.; Martin, J.; Mills, J.; Navani, N.; et al. British Thoracic Society guideline for diagnostic flexible bronchoscopy in adults: Accredited by NICE. Thorax 2013, 68, i1–i44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, W.-L.; Yang, J.; Strulovici-Barel, Y.; Salit, J.; Rostami, M.; Mezey, J.G.; O’Beirne, S.L.; Kaner, R.J.; Crystal, R.G. Exaggerated BMP4 signalling alters human airway basal progenitor cell differentiation to cigarette smoking-related phenotypes. Eur. Respir. J. 2019, 53, 1702553. [Google Scholar] [CrossRef] [PubMed]
- Perotin, J.-M.; Coraux, C.; Lagonotte, E.; Birembaut, P.; Delepine, G.; Polette, M.; Deslée, G.; Dormoy, V. Alteration of primary cilia in COPD. Eur. Respir. J. 2018, 52, 1800122. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diabasana, Z.; Perotin, J.-M.; Belgacemi, R.; Ancel, J.; Mulette, P.; Delepine, G.; Gosset, P.; Maskos, U.; Polette, M.; Deslée, G.; et al. Nicotinic Receptor Subunits Atlas in the Adult Human Lung. Int. J. Mol. Sci. 2020, 21, 7446. https://doi.org/10.3390/ijms21207446
Diabasana Z, Perotin J-M, Belgacemi R, Ancel J, Mulette P, Delepine G, Gosset P, Maskos U, Polette M, Deslée G, et al. Nicotinic Receptor Subunits Atlas in the Adult Human Lung. International Journal of Molecular Sciences. 2020; 21(20):7446. https://doi.org/10.3390/ijms21207446
Chicago/Turabian StyleDiabasana, Zania, Jeanne-Marie Perotin, Randa Belgacemi, Julien Ancel, Pauline Mulette, Gonzague Delepine, Philippe Gosset, Uwe Maskos, Myriam Polette, Gaëtan Deslée, and et al. 2020. "Nicotinic Receptor Subunits Atlas in the Adult Human Lung" International Journal of Molecular Sciences 21, no. 20: 7446. https://doi.org/10.3390/ijms21207446
APA StyleDiabasana, Z., Perotin, J. -M., Belgacemi, R., Ancel, J., Mulette, P., Delepine, G., Gosset, P., Maskos, U., Polette, M., Deslée, G., & Dormoy, V. (2020). Nicotinic Receptor Subunits Atlas in the Adult Human Lung. International Journal of Molecular Sciences, 21(20), 7446. https://doi.org/10.3390/ijms21207446