Protein Corona Gold Nanoparticles Fingerprinting Reveals a Profile of Blood Coagulation Proteins in the Serum of HER2-Overexpressing Breast Cancer Patients
Abstract
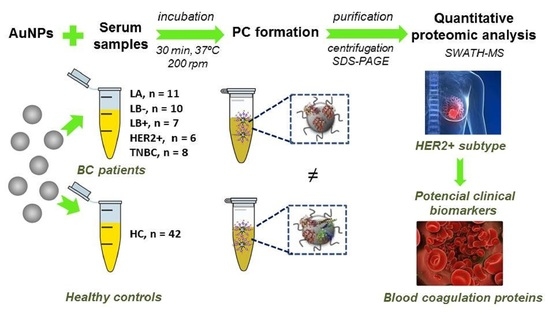
:1. Introduction
2. Results
2.1. Incubation of AuNPs (12.96 ± 0.72 nm) with Human Serum Samples: Ex Vivo Protein Corona Formation and Characterization
2.2. Quantitative Analysis of the Protein Corona-Coated AuNPs by SWATH-MS
2.3. Functional Pathway and Network Analysis for Subtype Specific Breast Cancer
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Biological Samples
4.3. Synthesis of Citrate-Gold Nanoparticles (AuNPs, 12.96 ± 0.72 nm)
4.4. Incubation of AuNPs with Human Serum Samples: Ex Vivo Protein Corona Formation
4.5. Characterization of Colloidal AuNPs
4.6. Separation and Digestion of the Proteins Presented in the Corona-Coated AuNPs
4.7. Protein Quantification by SWATH-MS
4.8. Protein Functional Interaction Network Analysis and Protein Ontology Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| APPs | Acute-phase proteins |
| AuNPs | Gold nanoparticles |
| BC | Breast cancer |
| DTT | Dithiothreitol |
| ER | Estrogen receptor |
| HC | Healthy controls |
| HER2 | Human epidermal growth factor receptor 2 |
| IAA | Iodoacetic acid |
| LA | Luminal A |
| LB | Luminal B |
| MS | Mass spectrometry |
| PC | Protein corona |
| PR | Progesterone receptor |
| SDS-PAGE | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SWATH-MS | Sequential Window Acquisition of All Theoretical Mass Spectra |
| TEM | Transmission electron microscopy |
| TNBC | Triple negative breast cancer |
| TF | Serotransferrin |
References
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef] [PubMed]
- Inic, Z.; Zegarac, M.; Inic, M.; Markovic, I.; Kozomara, Z.; Djurisic, I.; Inic, I.; Pupic, G.; Jancic, S. Difference between Luminal A and Luminal B Subtypes According to Ki-67, Tumor Size, and Progesterone Receptor Negativity Providing Prognostic Information. Clin. Med. Insights Oncol. 2014, 8, 107–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colleoni, M.; Sun, Z.; Price, K.N.; Karlsson, P.; Forbes, J.F.; Thürlimann, B.; Gianni, L.; Castiglione, M.; Gelber, R.D.; Coates, A.S.; et al. Annual Hazard Rates of Recurrence for Breast Cancer During 24 Years of Follow-Up: Results from the International Breast Cancer Study Group Trials I to V. J. Clin. Oncol. 2016, 34, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Asif, H.; Sultana, S.; Ahmed, S.; Akhtar, N.; Tariq, M. HER-2 Positive Breast Cance—A Mini-Review. Asian Pac. J. Cancer Prev. 2016, 17, 1609–1615. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Suman, S.; Basak, T.; Gupta, P.; Mishra, S.; Kumar, V.; Sengupta, S.; Shukla, Y. Quantitative Proteomics revealed novel proteins associated with molecular subtypes of breast cancer. J. Proteom. 2016, 148, 183–193. [Google Scholar] [CrossRef]
- Gajbhiye, A.; Dabhi, R.; Taunk, K.; Jagadeeshaprasad, M.G.; RoyChoudhury, S.; Mane, A.; Bayatigeri, S.; Chaudhury, K.; Santra, M.K.; Rapole, S. Multipronged quantitative proteomics reveals serum proteome alterations in breast cancer intrinsic subtypes. J. Proteom. 2017, 163, 1–13. [Google Scholar] [CrossRef]
- Bouchal, P.; Schubert, O.T.; Faktor, J.; Capkova, L.; Imrichova, H.; Zoufalova, K.; Paralova, V.; Hrstka, R.; Liu, Y.; Ebhardt, H.A.; et al. Breast Cancer Classification Based on Proteotypes Obtained by SWATH Mass Spectrometry. Cell Rep. 2019, 28, 832–843. [Google Scholar] [CrossRef]
- Jia, L.; Lu, Y.; Shao, J.; Liang, X.J.; Xu, Y. Nanoproteomics: A new sprout from emerging links between nanotechnology and proteomics. Trends Biotechnol. 2013, 31, 99–107. [Google Scholar] [CrossRef]
- Pozzi, D.; Caracciolo, G.; Digiacomo, L.; Colapicchioni, V.; Palchetti, S.; Capriotti, A.L.; Cavaliere, C.; Zenezini Chiozzi, R.; Puglisi, A.; Laganà, A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale 2015, 7, 13958–13966. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.P.; Castro López, A.; Bravo, S.B.; Vázquez-Estévez, S.; Acea-Nebril, B.; Núñez, C. Proteomic analysis of the bio-corona formed on the surface of (Au, Ag, Pt)-nanoparticles in human serum. Colloids Surf. B Biointerfaces 2019, 177, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.P.; Castro López, A.; García Vence, M.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials (Basel) 2020, 10, 1223. [Google Scholar] [CrossRef] [PubMed]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.K.; Stauber, R.H. The nanoparticle biomolecule corona: Lessons learned–challenge accepted? Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.W.; Yan, Y.; Caruso, F.; Nice, E.C. Emerging techniques in proteomics for probing nano-bio interactions. ACS Nano 2012, 6, 10438–10448. [Google Scholar] [CrossRef]
- Bai, X.; Wang, Y.; Song, Z.; Feng, Y.; Chen, Y.; Zhang, D.; Feng, L. The Basic Properties of Gold Nanoparticles and their Applications in Tumor Diagnosis and Treatment. Int. J. Mol. Sci. 2020, 21, 2480. [Google Scholar] [CrossRef] [Green Version]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles. The effect of size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [Green Version]
- Coussens, L.M.; Raymond, W.W.; Bergers, G. Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes Dev. 1999, 13, 1382–1397. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Berry, J.A.; Shoher, A.; Lucci, A. COX-2 induces IL-11 production in human breast cancer cells. J. Surg. Res. 2006, 131, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Singh, V.P.; Udupa, K.N. E-Rosette forming lymphocytes and serum immunoglobulins in breast cancer patients. Mater. Med. Pol. 1991, 23, 179–181. [Google Scholar] [PubMed]
- Lu, Y.; Hu, X. C5a stimulates the proliferation of breast cancer cells via Akt-dependent RGC-32 gene activation. Oncol. Rep. 2014, 32, 2817–2823. [Google Scholar] [CrossRef] [PubMed]
- Nasim, F.; Ejaz, S.; Ashraf, M.; Asif, A.R.; Oellerich, M.; Ahmad, G.; Malik, G.A.; Rehman, A.-U. Potential Biomarkers in the Sera of Breast Cancer Patients from Bahawalpur, Pakistan. Biomark. Cancer 2012, 4, 19–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.; Qu, H.; Yu, C.; Liu, J.; Jiao, A.; Sun, P. Serum biomarkers for lymph node metastasis in patients with triple-negative breast cancer by proteomics. Int. J. Clin. Exp. Pathol. 2017, 10, 3377–3384. [Google Scholar]
- Tiedemann, K.; Sadvakassova, G.; Mikolajewicz, N.; Juhas, M.; Sabirova, Z.; Tabariès, S.; Gettemans, J.; Siegel, P.M.; Komarova, S.V. Exosomal Release of L-Plastin by Breast Cancer Cells Facilitates Metastatic Bone Osteolysis. Transl. Oncol. 2019, 12, 462–474. [Google Scholar] [CrossRef]
- Ishikawa, S.; Miyashita, T.; Inokuchi, M.; Hayashi, H.; Oyama, K.; Tajima, H.; Takamura, H.; Ninomiya, I.; Ahmed, A.K.; Harman, J.W.; et al. Platelets surrounding primary tumor cells are related to chemoresistance. Oncol. Rep. 2016, 36, 787. [Google Scholar] [CrossRef] [Green Version]
- Goodarzi, A.; Yari, F.; Mohammadipour, M.; Deyhim, M.R.; Naghadeh, H.T. Capability of Platelet Factor 4 to Induce Apoptosis in the Cancerous Cell Lines in Vitro. Int. J. Med. Lab. 2018, 5, 195–207. [Google Scholar]
- Pendharkar, N.; Gajbhiye, A.; Taunk, K.; RoyChoudhury, S.; Dhali, S.; Seal, S.; Mane, A.; Abhang, S.; Santra, M.K.; Chaudhury, K.; et al. Quantitative tissue proteomic investigation of invasive ductal carcinoma of breast with luminal B HER2 positive and HER2 enriched subtypes towards potential diagnostic and therapeutic biomarkers. J. Proteome 2016, 132, 112–130. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, H.; Yang, P. Recent developments of nanoparticle-based enrichment methods for mass spectrometric analysis in proteomic. Sci. China Chem. 2010, 53, 695–703. [Google Scholar] [CrossRef]
- Asegaonkar, S.B.; Takalkar, U.V.; Kodlikeri, P.; Pagdhune, A.; Bonduliya, V.; Thorat, A.P. Serum high sensitivity C-reactive protein in breast cancer patients. Int. J. Res. Med. Sci. 2014, 2, 1408–1411. [Google Scholar] [CrossRef]
- Kaur, R.P.; Rubal; Banipal, R.P.S.; Vashistha, R.; Dhiman, M.; Munshi, A. Association of elevated levels of C-reactive protein with breast cancer, breast cancer subtypes and poor outcome. Curr. Probl. Cancer 2019, 43, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sun, X.; Lv, H.; Yang, X.; Kang, X. Serum amyloid A: A new potential serum marker correlated with the stage of breast cancer. Oncol. Lett. 2012, 3, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Ponzetti, M.; Capulli, M.; Angelucci, A.; Ventura, L.; Monache, S.D.; Mercurio, C.; Calgani, A.; Sanita, P.; Teti, A.; Rucci, N. Non-conventional role of haemoglobin beta in breast malignancy. Br. J. Cancer 2017, 117, 994–1006. [Google Scholar] [CrossRef] [Green Version]
- Vandewalle, B.; Hornez, L.; Revillion, F.; Lefebvre, J. Secretion of transferrin by human breast cancer cells. Biochem. Biophys. Res. Commun. 1989, 163, 149–154. [Google Scholar] [CrossRef]
- Lima, L.G.; Monteiro, R.Q. Activation of blood coagulation in cancer: Implications for tumour progression. Biosci. Rep. 2013, 33, e00064. [Google Scholar] [CrossRef]
- Metelli, A.; Wu, B.X.; Riesenberg, B.; Guglietta, S.; Huck, J.D.; Mills, C.; Li, A.; Rachidi, S.; Krieg, C.; Rubinstein, M.P.; et al. Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-β. Sci. Transl. Med. 2020, 12, eaay4860. [Google Scholar] [CrossRef]
- Hisada, Y.; Mackman, N. Cancer-associated pathways and biomarkers of venous thrombosis. Blood 2017, 130, 1499–1506. [Google Scholar] [CrossRef]
- Ruf, W.; Rothmeier, A.S.; Graf, C. Targeting clotting proteins in cancer therapy—Progress and challenges. Thromb. Res. 2016, 140, S1–S7. [Google Scholar] [CrossRef] [Green Version]
- Lal, I.; Dittus, K.; Holmes, C.E. Platelets, coagulation and fibrinolysis in breast cancer progression. Breast Cancer Res. 2013, 15, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tas, F.; Kilic, L.; Duranyildiz, D. Coagulation tests show significant differences in patients with breast cancer. Tumor Biol. 2014, 35, 5985–5992. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Lu, Q.; Liang, J.-W.; Xia, Y.; Zhang, W.; Hu, B.-Q.; Shang, F.-F.; Ji, Y.-R.; Wang, J.; Wang, Q.; et al. High Plasma Fibrinogen is Correlated with Poor Response to Trastuzumab Treatment in HER2 Positive Breast Cancer. Medicine (Baltimore) 2015, 94, e481. [Google Scholar] [CrossRef] [PubMed]
- Yigit, E.; Gönüllü, G.; Yücel, I.; Turgut, M.; Erdem, D.; Cakar, B. Relation between hemostatic parameters and prognostic/predictive factors in breast cancer. Eur. J. Intern. Med. 2008, 19, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Tinholt, M.; Garred, Ø.; Borgen, E.; Beraki, E.; Sletten, M.; Kleivi Sahlberg, K.; Sandset, P.M.; Iversen, N. Coagulation factor V is expressed in tumors and predicts favorable outcome in aggressive breast cancer. Thromb. Res. 2011, 164, S183. [Google Scholar] [CrossRef]
- Mutch, N.J. Emerging roles for factor XII in vivo. J. Thromb. Haemost. 2011, 9, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Kryza, T.; Silva, M.L.; Loessner, D.; Heuzé-Vourc’h, N.; Clements, J.A. The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie 2016, 122, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Zelvyte, I.; Sjögren, H.O.; Janciauskiene, S. Effects of native and cleaved forms of alpha1-antitrypsin on ME 1477 tumor cell functional activity. Cancer Detect. Prev. 2002, 26, 256–265. [Google Scholar] [CrossRef]
- Chan, H.J.; Li, H.; Liu, Z.; Yuan, Y.C.; Mortimer, J.; Chen, S. SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget 2015, 6, 25815–25827. [Google Scholar] [CrossRef] [Green Version]
- Luengo-Gil, G.; Calvo, M.I.; Martín-Villar, E.; Águila, S.; Bohdan, N.; Antón, A.I.; Espín, S.; de la Peña, F.A.; Vicente, V.; Corral, J.; et al. Antithrombin controls tumor migration, invasion and angiogenesis by inhibition of enteropeptidase. Sci. Rep. 2016, 6, 27544. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Cao, C.; Fu, J.; Li, Q.; Li, D.-H.; Chen, M.-Y. Serum adiponectin in breast cancer. A meta-analysis. Medicine (Baltimore) 2018, 97, e11433. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.W.; Park, C.-Y.; Lee, E.S.; Yoon, Y.S.; Lee, E.S.; Park, S.S.; Kim, Y.; Sung, N.J.; Yun, Y.H.; Lee, K.S.; et al. Adipokines, insulin resistance, metabolic syndrome, and breast cancer recurrence: A cohort study. Breast Cancer Res. 2011, 13, R34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Luo, G. Apolipoproteins, as the carrier proteins for lipids, are involved in the development of breast cancer. Clin. Transl. Oncol. 2020, 22, 1952–1962. [Google Scholar] [CrossRef] [PubMed]
- Cibeira, G.H.; Giacomazzi, J.; Aguiar, E.; Schneider, S.; Ettrich, B.; De Souza, C.I.; Camey, S.; Caleffi, M.; Weber, B.; Ashton-Prolla, P.; et al. Apolipoprotein E genetic polymorphism, serum lipoprotein levels and breast cancer risk: A case-control study. Mol. Clin. Oncol. 2014, 2, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-X.; Yuan, Q.; Min, Y.-L.; He, Y.; Xu, Q.-H.; Li, B.; Shi, W.-Q.; Lin, Q.; Li, Q.-H.; Zhu, P.-W.; et al. Apolipoprotein A1 and B as risk factors for development of intraocular metastasis in patients with breast cancer. Cancer Manag. Res. 2019, 11, 2881–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy 2012, 8, 1643–1656. [Google Scholar] [CrossRef] [Green Version]
- Whitelock, J.M.; Graham, L.D.; Melrose, J.; Murdoch, A.D.; Iozzo, R.V.; Underwood, P.A. Human perlecan immunopurified from different endothelial cell sources has different adhesive properties for vascular cells. Matrix Biol. 1999, 18, 163–178. [Google Scholar] [CrossRef]
- Kalscheuer, S.; Khanna, V.; Kim, H.; Li, S.; Sachdev, D.; DeCarlo, A.; Yang, D.; Panyam, J. Discovery of HSPG2 (Perlecan) as a Therapeutic Target in Triple Negative Breast Cancer. Sci. Rep. 2019, 9, 12492. [Google Scholar] [CrossRef]
- Davidson, B.; Stavnes, H.T.; Holth, A.; Chen, X.; Yang, Y.; Shih, I.-M.; Wang, T.-L. Gene expression signatures differentiate ovarian/peritoneal serous carcinoma from breast carcinoma in effusions. J. Cell. Mol. Med. 2011, 15, 535–544. [Google Scholar] [CrossRef]
- Aran, G.; Sanjurjo, L.; Bárcena, C.; Simon-Coma, M.; Téllez, E.; Vázquez-Vitali, M.; Garrido, M.; Guerra, L.; Díaz, E.; Ojanguren, I.; et al. CD5L is upregulated in hepatocellular carcinoma and promotes liver cancer cell proliferation and antiapoptotic responses by binding to HSPA5 (GRP78). FASEB J. 2018, 32, 3878–3891. [Google Scholar] [CrossRef] [Green Version]
- Dennison, J.B.; Molina, J.R.; Mitra, S.; González-Angulo, A.M.; Balko, J.M.; Kuba, M.G.; Sanders, M.E.; Pinto, J.A.; Gómez, H.L.; Arteaga, C.L.; et al. Lactate Dehydrogenase B: A Metabolic Marker of Response to Neoadjuvant Chemotherapy in Breast Cancer. Clin. Cancer Res. 2013, 19, 3703–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, E.; Araújo, J.E.; Gómez-Meire, S.; Lodeiro, C.; Pérez-Melón, C.; Iglesias-Lamas, E.; Otero-Glez, A.; Capelo, J.L.; Santos, H.M. Proteomics analysis of the peritoneal dialysate effluent reveals the presence of calcium-regulation proteins and acute inflammatory response. Clin. Proteom. 2014, 11, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]






| SWATH-MS Analysis | ||||||
|---|---|---|---|---|---|---|
| Comparison | Protein Number (p-Value ≤ 0.05) | |||||
| Total | Upregulated | Downregulated | Specific | Upregulated | Downregulated | |
| Controls vs. LA | 60 | 42 | 18 | 8 | 4 | 4 |
| Controls vs. LB- | 132 | 100 | 32 | 27 | 25 | 2 |
| Controls vs. LB+ | 67 | 59 | 8 | 2 | 2 | 0 |
| Controls vs. HER2+ | 130 | 95 | 35 | 28 | 23 | 5 |
| Controls vs. TNBC | 91 | 87 | 4 | 10 | 9 | 1 |
| Protein Name | Gene | p-Value | Fold Change | |
|---|---|---|---|---|
| Complement C1r subcomponent-like protein | C1RL | 0.0000979 | 1.614689351 | ↑ Luminal A |
| Complement factor H-related protein 2 | CFHR2 | 0.003228805 | 1.764346734 | ↑ Luminal A |
| Complement component C8 beta chain | C8B | 0.003730112 | 1.35440489 | ↑ Luminal A |
| Lysosome-associated membrane glycoprotein 2 | LAMP2 | 0.018383379 | 1.33466653 | ↑ Luminal A |
| Immunoglobulin kappa variable 3–20 | IGKV3–20 | 0.008581213 | 7.24153822 | ↓ Luminal A |
| Immunoglobulin heavy constant mu | IGHM | 0.038320909 | 2.180105502 | ↓ Luminal A |
| Immunoglobulin heavy variable 1–24 | IGHV1–24 | 0.045225189 | 2.766559939 | ↓ Luminal A |
| Protein Z-dependent protease inhibitor | SERPINA10 | 0.045960336 | 2.020513374 | ↓ Luminal A |
| Immunoglobulin lambda variable 2–23 | IGLV2–23 | 0.000000251 | 2.96506068 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy variable 3–53 | IGHV3–53 | 0.00000690 | 3.231686892 | ↑ Luminal B HER2 Neg |
| Immunoglobulin kappa variable 4–1 | IGKV4–1 | 0.00000788 | 2.271628981 | ↑ Luminal B HER2 Neg |
| Biotinidase | BTD | 0.0000142 | 1.571319968 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy constant alpha 1 | IGHA1 | 0.0000181 | 2.45029046 | ↑ Luminal B HER2 Neg |
| Serum paraoxonase/lactonase 3 | PON3 | 0.0000344 | 1.614088523 | ↑ Luminal B HER2 Neg |
| Immunoglobulin kappa constant | IGKC | 0.0000604 | 2.234283878 | ↑ Luminal B HER2 Neg |
| Phospholipid transfer protein | PLTP | 0.000127276 | 1.491697089 | ↑ Luminal B HER2 Neg |
| Immunoglobulin kappa variable 3–11 | IGKV3–11 | 0.000235484 | 2.727555354 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy variable 3–9 | IGHV3–9 | 0.000418498 | 2.709287474 | ↑ Luminal B HER2 Neg |
| Alpha-mannosidase 2 | MAN2A1 | 0.000577191 | 2.094925838 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy constant gamma 1 | IGHG1 | 0.000838666 | 1.832731289 | ↑ Luminal B HER2 Neg |
| Apolipoprotein B-100 | APOB | 0.001926757 | 1.618943103 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy constant alpha 2 | IGHA2 | 0.00207989 | 2.137794634 | ↑ Luminal B HER2 Neg |
| Basement membrane-specific heparan sulfate proteoglycan core protein | HSPG2 | 0.002185804 | 2.617271104 | ↑ Luminal B HER2 Neg |
| Pregnancy zone protein | PZP | 0.002663618 | 4.343426266 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy variable 1–69D | IGHV1–69D | 0.00307313 | 2.762019962 | ↑ Luminal B HER2 Neg |
| Immunoglobulin heavy variable 3–74 | IGHV3–74 | 0.005843078 | 1.63300774 | ↑ Luminal B HER2 Neg |
| Immunoglobulin lambda-like polypeptide 5 | IGLL5 | 0.006474796 | 1.674164832 | ↑ Luminal B HER2 Neg |
| Immunoglobulin kappa variable 3D-20 | IGKV3D-20 | 0.007387257 | 1.722873358 | ↑ Luminal B HER2 Neg |
| L-lactate dehydrogenase B chain | LDHB | 0.011088233 | 1.464826821 | ↑ Luminal B HER2 Neg |
| Platelet glycoprotein Ib alpha chain | GP1BA | 0.014355443 | 1.435497779 | ↑ Luminal B HER2 Neg |
| Apolipoprotein D | APOD | 0.02689663 | 1.565894604 | ↑ Luminal B HER2 Neg |
| Immunoglobulin lambda variable 3–21 | IGLV3–21 | 0.030887039 | 1.531598796 | ↑ Luminal B HER2 Neg |
| Mediator of RNA polymerase II transcription subunit 23 | MED23 | 0.034189832 | 1.582094144 | ↑ Luminal B HER2 Neg |
| Platelet factor 4 variant | PF4V1 | 0.013149247 | 8.695958011 | ↓ Luminal B HER2 Neg |
| Complement C1s subcomponent | C1S | 0.040782815 | 1.400513823 | ↓ Luminal B HER2 Neg |
| Plastin-2 | LCP1 | 0.002018957 | 4.501473239 | ↑ Luminal B HER2 Pos |
| Immunoglobulin heavy variable 6–1 | IGHV6–1 | 0.020191442 | 1.539338405 | ↑ Luminal B HER2 Pos |
| Complement C5 | C5 | 0.00000000000000295 | 2.104078338 | ↑ HER2 Pos |
| Adiponectin | ADIPOQ | 0.00000000828 | 9.579128591 | ↑ HER2 Pos |
| Immunoglobulin heavy variable 3–73 | IGHV3–73 | 0.00000167 | 15.87006684 | ↑ HER2 Pos |
| Coagulation factor XII | F12 | 0.00000250 | 4.48323521 | ↑ HER2 Pos |
| Plasma kallikrein | KLKB1 | 0.0000205 | 2.839283512 | ↑ HER2 Pos |
| Immunoglobulin heavy variable 3–23 | IGHV3–23 | 0.0000883 | 2.952483084 | ↑ HER2 Pos |
| Immunoglobulin lambda variable 1–51 | IGLV1–51 | 0.000382645 | 2.419267666 | ↑ HER2 Pos |
| Immunoglobulin heavy variable 3–64 | IGHV3–64 | 0.000401774 | 2.217759109 | ↑ HER2 Pos |
| Selenoprotein P | SELENOP | 0.000539614 | 3.796988329 | ↑ HER2 Pos |
| Immunoglobulin kappa variable 1D-12 | IGKV1D-12 | 0.00188481 | 4.471370936 | ↑ HER2 Pos |
| Immunoglobulin lambda variable 5–45 | IGLV5–45 | 0.00772078 | 2.858034161 | ↑ HER2 Pos |
| Immunoglobulin lambda variable 6–57 | IGLV6–57 | 0.009251968 | 5.252644969 | ↑ HER2 Pos |
| Keratin type I cytoskeletal 10 | KRT10 | 0.012361191 | 1.599012598 | ↑ HER2 Pos |
| Immunoglobulin kappa variable 1–27 | IGKV1–27 | 0.014636279 | 3.663302525 | ↑ HER2 Pos |
| Immunoglobulin kappa variable 1–5 | IGKV1–5 | 0.015089737 | 3.097611266 | ↑ HER2 Pos |
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 0.015909032 | 1.962284999 | ↑ HER2 Pos |
| Immunoglobulin kappa variable 2–24 | IGKV2–24 | 0.019946847 | 3.792528291 | ↑ HER2 Pos |
| Immunoglobulin heavy constant gamma 2 | IGHG2 | 0.020013155 | 1.605775681 | ↑ HER2 Pos |
| Adipocyte plasma membrane-associated protein | APMAP | 0.021204299 | 23.89017843 | ↑ HER2 Pos |
| Immunoglobulin kappa variable 1D-16 | IGKV1D-16 | 0.023927557 | 15.36602613 | ↑ HER2 Pos |
| Coagulation factor V | F5 | 0.025887503 | 3.739380413 | ↑ HER2 Pos |
| Cysteine-rich secretory protein 3 | CRISP3 | 0.034870563 | 3.338300324 | ↑ HER2 Pos |
| Immunoglobulin heavy variable 3–33 | IGHV3–33 | 0.038394512 | 8.344444793 | ↑ HER2 Pos |
| N-acetylmuramoyl-L-alanine amidase | PGLYRP2 | 0.000321687 | 1.799194526 | ↓ HER2 Pos |
| Alpha-1-antitrypsin | SERPINA1 | 0.001111283 | 4.959761437 | ↓ HER2 Pos |
| Trypsin-1 | PRSS1 | 0.002454431 | 4.217786063 | ↓ HER2 Pos |
| Apolipoprotein F | APOF | 0.005336626 | 7.55893037 | ↓ HER2 Pos |
| Antithrombin-III | SERPINC1 | 0.018612975 | 1.308597079 | ↓ HER2 Pos |
| Apolipoprotein E | APOE | 0.005108321 | 1.297484301 | ↑ Triple Negative |
| Voltage-dependent L-type calcium channel subunit alpha-1F | CACNA1F | 0.010760078 | 3.330169664 | ↑ Triple Negative |
| Complement C2 | C2 | 0.024115534 | 1.265184448 | ↑ Triple Negative |
| Keratin. type II cytoskeletal 1 | KRT1 | 0.02492244 | 1.394646766 | ↑ Triple Negative |
| Immunoglobulin heavy variable 4–30-2 | IGHV4–30-2 | 0.028590349 | 4.459516925 | ↑ Triple Negative |
| Attractin | ATRN | 0.033317422 | 1.248304377 | ↑ Triple Negative |
| Immunoglobulin kappa variable 2D-30 | IGKV2D-30 | 0.035772725 | 1.512873113 | ↑ Triple Negative |
| Immunoglobulin kappa variable 1–6 | IGKV1–6 | 0.039260496 | 1.537751007 | ↑ Triple Negative |
| Platelet basic protein | PPBP | 0.04971791 | 23.75806076 | ↑ Triple Negative |
| CD5 antigen-like | CD5L | 0.012543111 | 1.999836008 | ↓ Triple Negative |

| Protein Name | Gene | p-Value | Fold Change | Control vs. HER2 Positive |
|---|---|---|---|---|
| Coagulation factor XII | F12 | 0.00000250 | 4.48323521 | ↑ HER2 Pos |
| Plasma kallikrein | KLKB1 | 0.0000205 | 2.839283512 | ↑ HER2 Pos |
| Coagulation factor V | F5 | 0.025887503 | 3.739380413 | ↑ HER2 Pos |
| Alpha-1-antitrypsin | SERPINA1 | 0.001111283 | 4.959761437 | ↓ HER2 Pos |
| Trypsin-1 | PRSS1 | 0.002454431 | 4.217786063 | ↓ HER2 Pos |
| Antithrombin-III | SERPINC1 | 0.018612975 | 1.308597079 | ↓ HER2 Pos |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chantada-Vázquez, M.d.P.; Castro López, A.; García-Vence, M.; Acea-Nebril, B.; Bravo, S.B.; Núñez, C. Protein Corona Gold Nanoparticles Fingerprinting Reveals a Profile of Blood Coagulation Proteins in the Serum of HER2-Overexpressing Breast Cancer Patients. Int. J. Mol. Sci. 2020, 21, 8449. https://doi.org/10.3390/ijms21228449
Chantada-Vázquez MdP, Castro López A, García-Vence M, Acea-Nebril B, Bravo SB, Núñez C. Protein Corona Gold Nanoparticles Fingerprinting Reveals a Profile of Blood Coagulation Proteins in the Serum of HER2-Overexpressing Breast Cancer Patients. International Journal of Molecular Sciences. 2020; 21(22):8449. https://doi.org/10.3390/ijms21228449
Chicago/Turabian StyleChantada-Vázquez, María del Pilar, Antonio Castro López, María García-Vence, Benigno Acea-Nebril, Susana B. Bravo, and Cristina Núñez. 2020. "Protein Corona Gold Nanoparticles Fingerprinting Reveals a Profile of Blood Coagulation Proteins in the Serum of HER2-Overexpressing Breast Cancer Patients" International Journal of Molecular Sciences 21, no. 22: 8449. https://doi.org/10.3390/ijms21228449
APA StyleChantada-Vázquez, M. d. P., Castro López, A., García-Vence, M., Acea-Nebril, B., Bravo, S. B., & Núñez, C. (2020). Protein Corona Gold Nanoparticles Fingerprinting Reveals a Profile of Blood Coagulation Proteins in the Serum of HER2-Overexpressing Breast Cancer Patients. International Journal of Molecular Sciences, 21(22), 8449. https://doi.org/10.3390/ijms21228449