Pseudomonas aeruginosa Biofilms
Abstract
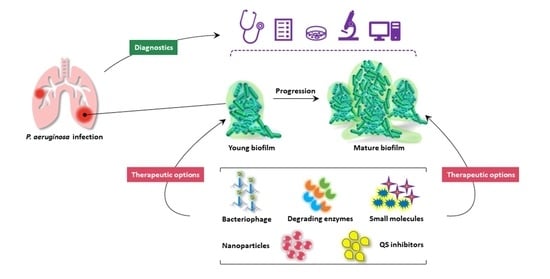
:1. Introduction
2. Pseudomonas aeruginosa Biofilm
2.1. Biofilm Composition
2.2. Biofilm Development
2.3. Multispecies Biofilm
3. Quorum Sensing in Biofilm Development
4. Diagnostics
4.1. Conventional Microbiological Culture
4.2. Molecular Biology Methods
4.3. Mass Spectrometry
4.4. Nanoparticle Biosensor
5. Therapeutic Strategies
5.1. Nanoparticles
5.2. Targeting EPS Component and Structure
5.3. Immunotherapies
5.4. Induction of Biofilm Dispersal
5.5. Inhibition of Quorum Sensing
5.6. Targeting Iron Metabolism
5.7. Photodynamic Therapy
5.8. Photothermal Therapy
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gale, M.J.; Maritato, M.S.; Chen, Y.-L.; Abdulateef, S.S.; Ruiz, J.E. Pseudomonas aeruginosa causing inflammatory mass of the nasopharynx in an immunocompromised HIV infected patient: A mimic of malignancy. IDCases 2015, 2, 40–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Jin, Y.; Bai, F.; Jin, S. Chapter 41—Pseudomonas Aeruginosa. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.-W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 753–767. [Google Scholar] [CrossRef]
- Gomila, A.; Carratalà, J.; Badia, J.M.; Camprubí, D.; Piriz, M.; Shaw, E.; Diaz-Brito, V.; Espejo, E.; Nicolás, C.; Brugués, M.; et al. Preoperative oral antibiotic prophylaxis reduces Pseudomonas aeruginosa surgical site infections after elective colorectal surgery: A multicenter prospective cohort study. BMC Infect. Dis. 2018, 18, 507. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K.; Gordillo, G.M.; Roy, S.; Kirsner, R.; Lambert, L.; Hunt, T.K.; Gottrup, F.; Gurtner, G.C.; Longaker, M.T. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair Regen 2009, 17, 763–771. [Google Scholar] [CrossRef] [Green Version]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Rollet, C.; Gal, L.; Guzzo, J. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: A comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 290, 135–142. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [Green Version]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef] [Green Version]
- Crespo, A.; Blanco-Cabra, N.; Torrents, E. Aerobic Vitamin B12 Biosynthesis Is Essential for Pseudomonas aeruginosa Class II Ribonucleotide Reductase Activity During Planktonic and Biofilm Growth. Front. Microbiol. 2018, 9, 986. [Google Scholar] [CrossRef] [PubMed]
- Oluyombo, O.; Penfold, C.N.; Diggle, S.P. Competition in Biofilms between Cystic Fibrosis Isolates of Pseudomonas aeruginosa Is Shaped by R-Pyocins. mBio 2019, 10, e01828-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coughlan, L.M.; Cotter, P.D.; Hill, C.; Alvarez-Ordonez, A. New Weapons to Fight Old Enemies: Novel Strategies for the (Bio)control of Bacterial Biofilms in the Food Industry. Front. Microbiol. 2016, 7, 1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moradali, M.F.; Rehm, B.H.A. Bacterial biopolymers: From pathogenesis to advanced materials. Nat. Rev. Microbiol. 2020, 18, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.H.A. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Strempel, N.; Neidig, A.; Nusser, M.; Geffers, R.; Vieillard, J.; Lesouhaitier, O.; Brenner-Weiss, G.; Overhage, J. Human host defense peptide LL-37 stimulates virulence factor production and adaptive resistance in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e82240. [Google Scholar] [CrossRef]
- Jackson, K.D.; Starkey, M.; Kremer, S.; Parsek, M.R.; Wozniak, D.J. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 2004, 186, 4466–4475. [Google Scholar] [CrossRef] [Green Version]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Billings, N.; Millan, M.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 2013, 9, e1003526. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Conover, M.; Lu, H.; Parsek, M.R.; Bayles, K.; Wozniak, D.J. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009, 5, e1000354. [Google Scholar] [CrossRef] [Green Version]
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Wang, S.; Wang, D.; Parsek, M.R.; Wozniak, D.J. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 2012, 65, 377–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, C.J.; Wozniak, D.J. Psl Produced by Mucoid Pseudomonas aeruginosa Contributes to the Establishment of Biofilms and Immune Evasion. mBio 2017, 8, e00864-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irie, Y.; Roberts, A.E.L.; Kragh, K.N.; Gordon, V.D.; Hutchison, J.; Allen, R.J.; Melaugh, G.; Bjarnsholt, T.; West, S.A.; Diggle, S.P. The Pseudomonas aeruginosa PSL Polysaccharide Is a Social but Noncheatable Trait in Biofilms. mBio 2017, 8, e00374-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staudinger, B.J.; Muller, J.F.; Halldórsson, S.; Boles, B.; Angermeyer, A.; Nguyen, D.; Rosen, H.; Baldursson, O.; Gottfreðsson, M.; Guðmundsson, G.H.; et al. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 2014, 189, 812–824. [Google Scholar] [CrossRef] [Green Version]
- Irie, Y.; Borlee, B.R.; O’Connor, J.R.; Hill, P.J.; Harwood, C.S.; Wozniak, D.J.; Parsek, M.R. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2012, 109, 20632–20636. [Google Scholar] [CrossRef] [Green Version]
- Mishra, M.; Byrd, M.S.; Sergeant, S.; Azad, A.K.; Parsek, M.R.; McPhail, L.; Schlesinger, L.S.; Wozniak, D.J. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012, 14, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Colvin, K.M.; Alnabelseya, N.; Baker, P.; Whitney, J.C.; Howell, P.L.; Parsek, M.R. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2329–2339. [Google Scholar] [CrossRef] [Green Version]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [Green Version]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690. [Google Scholar] [CrossRef]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2011, 14, 1913–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Hu, Y.; Liu, Y.; Zhang, J.; Ulstrup, J.; Molin, S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 2011, 13, 1705–1717. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.; Hill, P.J.; Snarr, B.D.; Alnabelseya, N.; Pestrak, M.J.; Lee, M.J.; Jennings, L.K.; Tam, J.; Melnyk, R.A.; Parsek, M.R.; et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016, 2, e1501632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciofu, O.; Tolker-Nielsen, T.; Jensen, P.Ø.; Wang, H.; Høiby, N. Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv. Drug Deliv. Rev. 2015, 85, 7–23. [Google Scholar] [CrossRef]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Genet. 2012, 10, 841–851. [Google Scholar] [CrossRef]
- Evans, L.R.; Linker, A. Production and Characterization of the Slime Polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973, 116, 915–924. [Google Scholar] [CrossRef] [Green Version]
- Tseng, B.S.; Zhang, W.; Harrison, J.J.; Quach, T.P.; Song, J.L.; Penterman, J.; Singh, P.K.; Chopp, D.L.; Packman, A.I.; Parsek, M.R. The extracellular matrix protectsPseudomonas aeruginosabiofilms by limiting the penetration of tobramycin. Environ. Microbiol. 2013, 15, 2865–2878. [Google Scholar] [CrossRef] [Green Version]
- Hay, I.D.; Rehman, Z.U.; Moradali, M.F.; Wang, Y.; Rehm, B.H.A. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013, 6, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Hay, I.D.; Rehman, Z.U.; Ghafoor, A.; Rehm, B.H.A. Bacterial biosynthesis of alginates. J. Chem. Technol. Biotechnol. 2010, 85, 752–759. [Google Scholar] [CrossRef]
- Gloag, E.S.; German, G.K.; Stoodley, P.; Wozniak, D.J. Viscoelastic properties of Pseudomonas aeruginosa variant biofilms. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wloka, M.; Rehage, H.; Flemming, H.-C.; Wingender, J. Structure and rheological behaviour of the extracellular polymeric substance network of mucoid Pseudomonas aeruginosa biofilms. Biofilms 2005, 2, 275–283. [Google Scholar] [CrossRef]
- Rehm, B.H.A.; Valla, S. Bacterial alginates: Biosynthesis and applications. Appl. Microbiol. Biotechnol. 1997, 48, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef] [Green Version]
- Chiang, W.-C.; Nilsson, M.; Jensen, P.Ø.; Høiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Extracellular DNA Shields against Aminoglycosides in Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2013, 57, 2352–2361. [Google Scholar] [CrossRef] [Green Version]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.C.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 60, 544–553. [Google Scholar] [CrossRef] [Green Version]
- Wilton, M.; Wong, M.J.Q.; Tang, L.; Liang, X.; Moore, R.; Parkins, M.D.; Lewenza, S.; Dong, T.G. Chelation of Membrane-Bound Cations by Extracellular DNA Activates the Type VI Secretion System in Pseudomonas aeruginosa. Infect. Immun. 2016, 84, 2355–2361. [Google Scholar] [CrossRef] [Green Version]
- Gloag, E.S.; Turnbull, L.; Huang, A.; Vallotton, P.; Wang, H.; Nolan, L.M.; Mililli, L.; Hunt, C.; Lu, J.; Osvath, S.R.; et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. USA 2013, 110, 11541–11546. [Google Scholar] [CrossRef] [Green Version]
- Bass, J.I.F.; Russo, D.M.; Gabelloni, M.L.; Geffner, J.R.; Giordano, M.; Catalano, M.; Zorreguieta, Á.; Trevani, A.S. Extracellular DNA: A Major Proinflammatory Component of Pseudomonas aeruginosaBiofilms. J. Immunol. 2010, 184, 6386–6395. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.H.; Webb, J.S.; Rehm, B.H.A. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology 2004, 150, 3405–3413. [Google Scholar] [CrossRef] [Green Version]
- Sønderholm, M.; Kragh, K.N.; Koren, K.; Jakobsen, T.H.; Darch, S.E.; Alhede, M.; Jensen, P.Ø.; Whiteley, M.; Kühl, M.; Bjarnsholt, T. Pseudomonas aeruginosa Aggregate Formation in an Alginate Bead Model System Exhibits In Vivo-Like Characteristics. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosabiofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003, 50, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Hickman, J.W.; Harwood, C.S. Identification of FleQ fromPseudomonas aeruginosaas a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008, 69, 376–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilbaud, M.; Bruzaud, J.; Bouffartigues, E.; Orange, N.; Guillot, A.; Aubert-Frambourg, A.; Monnet, V.; Herry, J.-M.; Chevalier, S.; Bellon-Fontaine, M.-N. Proteomic Response of Pseudomonas aeruginosa PAO1 Adhering to Solid Surfaces. Front. Microbiol. 2017, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [Green Version]
- Cherny, K.E.; Sauer, K. Pseudomonas aeruginosa Requires the DNA-Specific Endonuclease EndA To Degrade Extracellular Genomic DNA To Disperse from the Biofilm. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef]
- Chua, S.L.; Tan, S.Y.-Y.; Rybtke, M.T.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Tolker-Nielsen, T.; Yang, L.; Givskov, M. Bis-(3′-5′)-Cyclic Dimeric GMP Regulates Antimicrobial Peptide Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 2066–2075. [Google Scholar] [CrossRef] [Green Version]
- Chua, S.L.; Hultqvist, L.D.; Yuan, M.; Rybtke, M.; E Nielsen, T.; Givskov, M.; Tolker-Nielsen, T.; Yang, L. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm–dispersed cells via c-di-GMP manipulation. Nat. Protoc. 2015, 10, 1165–1180. [Google Scholar] [CrossRef]
- Chua, S.L.; Liu, Y.; Yam, J.K.H.; Chen, Y.; Vejborg, R.M.; Tan, B.G.C.; Kjelleberg, S.; Tolker-Nielsen, T.; Givskov, M.; Yang, L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 2014, 5, 4462. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.R.; Cherny, K.E.; Sauer, K. Susceptibility of Pseudomonas aeruginosa Dispersed Cells to Antimicrobial Agents Is Dependent on the Dispersion Cue and Class of the Antimicrobial Agent Used. Antimicrob. Agents Chemother. 2017, 61, e00846-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Petrova, O.E.; Su, S.; Lau, G.W.; Panmanee, W.; Na, R.; Hassett, D.J.; Davies, D.G.; Sauer, K. BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1004168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Mashburn, L.M.; Jett, A.M.; Akins, D.R.; Whiteley, M. Staphylococcus aureus Serves as an Iron Source for Pseudomonas aeruginosa during In Vivo Coculture. J. Bacteriol. 2005, 187, 554–566. [Google Scholar] [CrossRef] [Green Version]
- Korgaonkar, A.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. USA 2012, 110, 1059–1064. [Google Scholar] [CrossRef] [Green Version]
- Willner, D.L.; Haynes, M.R.; Furlan, M.; Schmieder, R.; Lim, Y.W.; Rainey, P.B.; Rohwer, F.; Conrad, D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012, 6, 471–474. [Google Scholar] [CrossRef] [Green Version]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Weaver, V.B.; Kolter, R. Burkholderia spp. Alter Pseudomonas aeruginosa Physiology through Iron Sequestration. J. Bacteriol. 2004, 186, 2376–2384. [Google Scholar] [CrossRef] [Green Version]
- Limoli, D.H.; Whitfield, G.B.; Kitao, T.; Ivey, M.L.; Davis, M.R., Jr.; Grahl, N.; Hogan, D.A.; Rahme, L.G.; Howell, P.L.; O’Toole, G.A.; et al. Pseudomonas aeruginosaAlginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. mBio 2017, 8, e00186-17. [Google Scholar] [CrossRef] [Green Version]
- Armbruster, C.R.; Wolter, D.J.; Mishra, M.; Hayden, H.S.; Radey, M.C.; Merrihew, G.; MacCoss, M.J.; Burns, J.; Wozniak, D.J.; Parsek, M.R.; et al. Staphylococcus aureus Protein A Mediates Interspecies Interactions at the Cell Surface of Pseudomonas aeruginosa. mBio 2016, 7, e00538-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, S.C.; Yam, J.K.H.; Matysik, A.; Seng, Z.J.; Klebensberger, J.; Givskov, M.; Doyle, P.; Rice, S.A.; Yang, L.; Kjelleberg, S. Matrix Polysaccharides and SiaD Diguanylate Cyclase Alter Community Structure and Competitiveness of Pseudomonas aeruginosa during Dual-Species Biofilm Development with Staphylococcus aureus. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragonzi, A.; Farulla, I.; Paroni, M.; Twomey, K.B.; Pirone, L.; Lorè, N.I.; Bianconi, I.; Dalmastri, C.; Ryan, R.P.; Bevivino, A. Modelling Co-Infection of the Cystic Fibrosis Lung by Pseudomonas aeruginosa and Burkholderia cenocepacia Reveals Influences on Biofilm Formation and Host Response. PLoS ONE 2012, 7, e52330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filkins, L.M.; Hampton, T.H.; Gifford, A.H.; Gross, M.J.; Hogan, D.A.; Sogin, M.L.; Morrison, H.G.; Paster, B.J.; O’Toole, G.A. Prevalence of Streptococci and Increased Polymicrobial Diversity Associated with Cystic Fibrosis Patient Stability. J. Bacteriol. 2012, 194, 4709–4717. [Google Scholar] [CrossRef] [Green Version]
- Scoffield, J.A.; Wu, H. Oral Streptococci and Nitrite-Mediated Interference of Pseudomonas aeruginosa. Infect. Immun. 2015, 83, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scoffield, J.A.; Duan, D.; Zhu, F.; Wu, H. A commensal streptococcus hijacks a Pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PLoS Pathog. 2017, 13, e1006300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Wu, G. Can Biofilm Be Reversed Through Quorum Sensing in Pseudomonas aeruginosa? Front. Microbiol. 2019, 10, 1582. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Genet. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Park, H.-Y.; Park, S.J.; Park, S.-J.; Kim, S.-K.; Ha, C.; Im, S.-J.; Lee, J.-H. Growth phase-differential quorum sensing regulation of anthranilate metabolism in Pseudomonas aeruginosa. Mol. Cells 2011, 32, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Oglesby, A.G.; Farrow, J.M.I.; Lee, J.-H.; Tomaras, A.P.; Greenberg, E.P.; Pesci, E.C.; Vasil, M.L. The Influence of Iron on Pseudomonas aeruginosa Physiology: A regulatory link between iron and quorum sensing. J. Biol. Chem. 2008, 283, 15558–15567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampioni, G.; Falcone, M.; Heeb, S.; Frangipani, E.; Fletcher, M.P.; Dubern, J.-F.; Visca, P.; Leoni, L.; Cámara, M.; Williams, P. Unravelling the Genome-Wide Contributions of Specific 2-Alkyl-4-Quinolones and PqsE to Quorum Sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016, 12, e1006029. [Google Scholar] [CrossRef] [PubMed]
- Mashburn, L.M.; Whiteley, M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nat. Cell Biol. 2005, 437, 422–425. [Google Scholar] [CrossRef] [PubMed]
- Seed, P.C.; Passador, L.; Iglewski, B.H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: An autoinduction regulatory hierarchy. J. Bacteriol. 1995, 177, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Déziel, E.; Lépine, F.; Milot, S.; He, J.; Mindrinos, M.N.; Tompkins, R.G.; Rahme, L.G. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2004, 101, 1339–1344. [Google Scholar] [CrossRef] [Green Version]
- Winson, M.K.; Camara, M.; Latifi, A.; Foglino, M.; Chhabra, S.R.; Daykin, M.; Bally, M.; Chapon, V.; Salmond, G.P.; Bycroft, B.W. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 9427–9431. [Google Scholar] [CrossRef] [Green Version]
- McKnight, S.L.; Iglewski, B.H.; Pesci, E.C. The Pseudomonas Quinolone Signal Regulates rhl Quorum Sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 2702–2708. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Krishnan, G.; Goumnerov, B.; Tsongalis, J.; Tompkins, R.; Rahme, L.G. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 2001, 98, 14613–14618. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef]
- Meng, X.; Ahator, S.D.; Zhang, L.-H. Molecular Mechanisms of Phosphate Stress Activation of Pseudomonas aeruginosa Quorum Sensing Systems. mSphere 2020, 5, 119–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, L.; Cornelis, P.; Guillemyn, K.; Ballet, S.; Christophersen, C.; Hammerich, O. Structure Revision of N-Mercapto-4-formylcarbostyril Produced by Pseudomonas fluorescens G308 to 2-(2-Hydroxyphenyl)thiazole-4-carbaldehyde [aeruginaldehyde]. Nat. Prod. Commun. 2014, 9, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Trottmann, F.; Franke, J.; Ishida, K.; Garcia-Altares, M.; Hertweck, C. A Pair of Bacterial Siderophores Releases and Traps an Intercellular Signal Molecule: An Unusual Case of Natural Nitrone Bioconjugation. Angew. Chem. Int. Ed. 2018, 58, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Murcia, N.E.; Elee, X.; Ewaridel, P.; Emaspoli, A.; Imker, H.J.; Echai, T.; Walsh, C.T.; Reimmann, C. The Pseudomonas aeruginosa antimetabolite L -2-amino-4-methoxy-trans-3-butenoic acid (AMB) is made from glutamate and two alanine residues via a thiotemplate-linked tripeptide precursor. Front. Microbiol. 2015, 6, 170. [Google Scholar] [CrossRef] [Green Version]
- Pamp, S.J.; Tolker-Nielsen, T. Multiple Roles of Biosurfactants in Structural Biofilm Development by Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2531–2539. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.P.; Pesci, E.C.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef] [Green Version]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. From The Cover: Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Kutty, S.K.; Kumar, N.; Manefield, M. Pyocyanin Facilitates Extracellular DNA Binding to Pseudomonas aeruginosa Influencing Cell Surface Properties and Aggregation. PLoS ONE 2013, 8, e58299. [Google Scholar] [CrossRef]
- Das, T.; Kutty, S.K.; Tavallaie, R.; Ibugo, A.I.; Panchompoo, J.; Sehar, S.; Aldous, L.; Yeung, A.W.S.; Thomas, S.R.; Kumar, N.; et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci. Rep. 2015, 5, srep08398. [Google Scholar] [CrossRef]
- Sakuragi, Y.; Kolter, R. Quorum-Sensing Regulation of the Biofilm Matrix Genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5383–5386. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, D.P.; Matwichuk, M.L.; Townsend, D.O.; Reichhardt, C.; Lamba, D.; Wozniak, D.J.; Parsek, M.R. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Diggle, S.P.; Stacey, R.E.; Dodd, C.; Cámara, M.; Williams, P.; Winzer, K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 2006, 8, 1095–1104. [Google Scholar] [CrossRef]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Kirisits, M.J.; Prost, L.; Starkey, M.; Parsek, M.R. Characterization of Colony Morphology Variants Isolated from Pseudomonas aeruginosa Biofilms. Appl. Environ. Microbiol. 2005, 71, 4809–4821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whistler, T.; Sangwichian, O.; Jorakate, P.; Sawatwong, P.; Surin, U.; Piralam, B.; Thamthitiwat, S.; Promkong, C.; Peruski, L. Identification of Gram negative non-fermentative bacteria: How hard can it be? PLoS Negl. Trop. Dis. 2019, 13, e0007729. [Google Scholar] [CrossRef] [PubMed]
- Abayasekara, L.M.; Perera, J.; Chandrasekharan, V.; Gnanam, V.S.; Udunuwara, N.A.; Liyanage, D.S.; Bulathsinhala, N.E.; Adikary, S.; Aluthmuhandiram, J.V.S.; Thanaseelan, C.S.; et al. Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: A comparative study. BMC Infect. Dis. 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bobenchik, A.M.; Deak, E.; Hindler, J.A.; Charlton, C.L.; Humphries, R.M. Performance of Vitek 2 for Antimicrobial Susceptibility Testing of Acinetobacter baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia with Vitek 2 (2009 FDA) and CLSI M100S 26th Edition Breakpoints. J. Clin. Microbiol. 2016, 55, 450–456. [Google Scholar] [CrossRef] [Green Version]
- Sader, H.S.; Fritsche, T.R.; Jones, R.N. Accuracy of Three Automated Systems (MicroScan WalkAway, VITEK, and VITEK 2) for Susceptibility Testing of Pseudomonas aeruginosa against Five Broad-Spectrum Beta-Lactam Agents. J. Clin. Microbiol. 2006, 44, 1101–1104. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, C.M. Evaluation of the Phoenix 100 ID/AST System and NID Panel for Identification of Enterobacteriaceae, Vibrionaceae, and Commonly Isolated Nonenteric Gram-Negative Bacilli. J. Clin. Microbiol. 2006, 44, 928–933. [Google Scholar] [CrossRef] [Green Version]
- McGregor, A.; Schio, F.; Beaton, S.; Boulton, V.; Perman, M.; Gilbert, G. The MicroScan WalkAway diagnostic microbiology system—An evaluation. Pathology 1995, 27, 172–176. [Google Scholar] [CrossRef]
- Ligozzi, M.; Bernini, C.; Bonora, M.G.; De Fatima, M.; Zuliani, J.; Fontana, R.J. Evaluation of the VITEK 2 System for Identification and Antimicrobial Susceptibility Testing of Medically Relevant Gram-Positive Cocci. J. Clin. Microbiol. 2002, 40, 1681–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekyere, J.O.; Sephofane, A.K.; Mbelle, N.M. Comparative Evaluation of CHROMagar COL-APSE, MicroScan Walkaway, ComASP Colistin, and Colistin MAC Test in Detecting Colistin-resistant Gram-Negative Bacteria. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Emerson, J.; Stapp, J.; Stapp, L.; Abe, P.; Burns, J.L. Use of Real-Time PCR with Multiple Targets To Identify Pseudomonas aeruginosa and Other Nonfermenting Gram-Negative Bacilli from Patients with Cystic Fibrosis. J. Clin. Microbiol. 2003, 41, 4312–4317. [Google Scholar] [CrossRef] [Green Version]
- Anuj, S.N.; Whiley, D.M.; Kidd, T.J.; Bell, S.C.; Wainwright, C.E.; Nissen, M.D.; Sloots, T.P. Identification of Pseudomonas aeruginosa by a duplex real-time polymerase chain reaction assay targeting the ecfX and the gyrB genes. Diagn. Microbiol. Infect. Dis. 2009, 63, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Aghamollaei, H.; Moghaddam, M.M.; Kooshki, H.; Heiat, M.; Mirnejad, R.; Barzi, N.S. Detection of Pseudomonas aeruginosa by a triplex polymerase chain reaction assay based on lasI/R and gyrB genes. J. Infect. Public Health 2015, 8, 314–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motoshima, M.; Yanagihara, K.; Fukushima, K.; Matsuda, J.; Sugahara, K.; Hirakata, Y.; Yamada, Y.; Kohno, S.; Kamihira, S. Rapid and accurate detection of Pseudomonas aeruginosa by real-time polymerase chain reaction with melting curve analysis targeting gyrB gene. Diagn. Microbiol. Infect. Dis. 2007, 58, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, F.; Le Berre, R.; Rosec, S.; Hardy, J.; Gouriou, S.; Boisramé-Gastrin, S.; Vallet, S.; Rault, G.; Payan, C.; Héry-Arnaud, G. Proposal of a quantitative PCR-based protocol for an optimal Pseudomonas aeruginosa detection in patients with cystic fibrosis. BMC Microbiol. 2013, 13, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edong, D.; Ezou, D.; Eliu, H.; Eyang, Z.; Ehuang, S.; Eliu, N.; Ehe, X.; Eliu, W.; Huang, L. Rapid detection of Pseudomonas aeruginosa targeting the toxA gene in intensive care unit patients from Beijing, China. Front. Microbiol. 2015, 6, 1100. [Google Scholar] [CrossRef] [Green Version]
- Oumeraci, T.; Jensen, V.; Talbot, S.R.; Hofmann, W.; Kostrzewa, M.; Schlegelberger, B.; Von Neuhoff, N.; Häussler, S. Comprehensive MALDI-TOF Biotyping of the Non-Redundant Harvard Pseudomonas aeruginosa PA14 Transposon Insertion Mutant Library. PLoS ONE 2015, 10, e0117144. [Google Scholar] [CrossRef] [Green Version]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Li, H.; Lu, X.; Stratton, C.W.; Tang, Y.-W. Mass Spectrometry Biotyper System Identifies Enteric Bacterial Pathogens Directly from Colonies Grown on Selective Stool Culture Media. J. Clin. Microbiol. 2010, 48, 3888–3892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonnelle, E.; Beretti, J.-L.; Cottyn, S.; Quesne, G.; Berche, P.; Nassif, X.; Ferroni, A. Rapid Identification of Staphylococci Isolated in Clinical Microbiology Laboratories by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2007, 45, 2156–2161. [Google Scholar] [CrossRef] [Green Version]
- Sogawa, K.; Watanabe, M.; Sato, K.; Segawa, S.; Ishii, C.; Miyabe, A.; Murata, S.; Saito, T.; Nomura, F. Use of the MALDI BioTyper system with MALDI–TOF mass spectrometry for rapid identification of microorganisms. Anal. Bioanal. Chem. 2011, 400, 1905–1911. [Google Scholar] [CrossRef]
- Cabrolier, N.; Sauget, M.; Bertrand, X.; Hocquet, D. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Identifies Pseudomonas aeruginosa High-Risk Clones. J. Clin. Microbiol. 2015, 53, 1395–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Treviño, S.; Garza-González, E.; Mendoza-Olazarán, S.; Morfín-Otero, R.; Camacho-Ortiz, A.; Rodríguez-Noriega, E.; Martínez-Meléndez, A.; Bocanegra-Ibarias, P. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, F.D.; Bonatto, C.C.; Lopes, C.A.; Pereira, A.L.; Silva, L.P. Use of MALDI-TOF mass spectrometry to analyze the molecular profile of Pseudomonas aeruginosa biofilms grown on glass and plastic surfaces. Microb. Pathog. 2015, 86, 32–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBritto, S.; Gajbar, T.D.; Satapute, P.; Sundaram, L.; Lakshmikantha, R.Y.; Jogaiah, S.; Ito, S.-I. Isolation and characterization of nutrient dependent pyocyanin from Pseudomonas aeruginosa and its dye and agrochemical properties. Sci. Rep. 2020, 10, 1542. [Google Scholar] [CrossRef] [Green Version]
- Barequet, I.S.; Ben Simon, G.J.; Safrin, M.; Ohman, D.E.; Kessler, E. Pseudomonas aeruginosa LasA Protease in Treatment of Experimental Staphylococcal Keratitis. Antimicrob. Agents Chemother. 2004, 48, 1681–1687. [Google Scholar] [CrossRef] [Green Version]
- Elkhawaga, A.A.; Khalifa, M.M.; El-Badawy, O.; Hassan, M.A.; El-Said, W.A. Rapid and highly sensitive detection of pyocyanin biomarker in different Pseudomonas aeruginosa infections using gold nanoparticles modified sensor. PLoS ONE 2019, 14, e0216438. [Google Scholar] [CrossRef] [Green Version]
- AlHogail, S.; Suaifan, G.A.; Bikker, F.J.; Kaman, W.E.; Weber, K.; Cialla-May, D.; Meyer, T.; Zourob, M. Rapid Colorimetric Detection of Pseudomonas aeruginosa in Clinical Isolates Using a Magnetic Nanoparticle Biosensor. ACS Omega 2019, 4, 21684–21688. [Google Scholar] [CrossRef] [Green Version]
- Breidenstein, E.B.; De La Fuente-Nunez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Yamada, K.M. Dynamic cell-matrix interactions modulate microbial biofilm and tissue 3D microenvironments. Curr. Opin. Cell Biol. 2016, 42, 102–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajishengallis, G.; Hajishengallis, E.; Kajikawa, T.; Wang, B.; Yancopoulou, D.; Ricklin, D.; Lambris, J.D. Complement inhibition in pre-clinical models of periodontitis and prospects for clinical application. Semin. Immunol. 2016, 28, 285–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watters, C.; Everett, J.A.; Haley, C.; Clinton, A.; Rumbaugh, K. Insulin Treatment Modulates the Host Immune System To Enhance Pseudomonas aeruginosa Wound Biofilms. Infect. Immun. 2013, 82, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Maliniak, M.; Stecenko, A.A.; Mccarty, N.A. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: A single-center study. J. Cyst. Fibros. 2016, 15, 350–356. [Google Scholar] [CrossRef] [Green Version]
- Reffuveille, F.; De La Fuente-Nunez, C.; Fairfull-Smith, K.E.; Hancock, R.E. Potentiation of ciprofloxacin action against Gram-negative bacterial biofilms by a nitroxide. Pathog. Dis. 2015, 73, 73. [Google Scholar] [CrossRef] [Green Version]
- Aoki, W.; Ueda, M. Characterization of Antimicrobial Peptides toward the Development of Novel Antibiotics. Pharmaceuticals 2013, 6, 1055–1081. [Google Scholar] [CrossRef] [Green Version]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A Review of Antimicrobial Peptides and Their Therapeutic Potential as Anti-Infective Drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef]
- Papareddy, P.; Kasetty, G.; Kalle, M.; Bhongir, R.K.V.; Mörgelin, M.; Schmidtchen, A.; Malmsten, M. NLF20: An antimicrobial peptide with therapeutic potential against invasive Pseudomonas aeruginosa infection. J. Antimicrob. Chemother. 2015, 71, 170–180. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.C.; Ahn, I.H.; Ahn, D.H.; Lee, Y.M.; Lee, J.; Lee, J.H.; Kim, H.-W.; Park, H. Characterization of Two Antimicrobial Peptides from Antarctic Fishes (Notothenia coriiceps and Parachaenichthys charcoti). PLoS ONE 2017, 12, e0170821. [Google Scholar] [CrossRef] [PubMed]
- Wnorowska, U.; Niemirowicz, K.; Myint, M.; Diamond, S.L.; Wróblewska, M.; Savage, P.B.; Janmey, P.A.; Bucki, R. Bactericidal Activities of Cathelicidin LL-37 and Select Cationic Lipids against the Hypervirulent Pseudomonas aeruginosa Strain LESB58. Antimicrob. Agents Chemother. 2015, 59, 3808–3815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flume, P.A.; VanDevanter, D.R.; Morgan, E.E.; Dudley, M.N.; Loutit, J.S.; Bell, S.C.; Kerem, E.; Fischer, R.; Smyth, A.R.; Aaron, S.D.; et al. A phase 3, multi-center, multinational, randomized, double-blind, placebo-controlled study to evaluate the efficacy and safety of levofloxacin inhalation solution (APT-1026) in stable cystic fibrosis patients. J. Cyst. Fibros. 2016, 15, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, S.; Hauck, D.; Hoffmann, M.; Sommer, R.; Joachim, I.; Müller, R.; Imberty, A.; Varrot, A.; Titz, A. Covalent Lectin Inhibition and Application in Bacterial Biofilm Imaging. Angew. Chem. Int. Ed. 2017, 56, 16559–16564. [Google Scholar] [CrossRef] [Green Version]
- Johansson, E.M.; Crusz, S.A.; Kolomiets, E.; Buts, L.; Kadam, R.U.; Cacciarini, M.; Bartels, K.-M.; Diggle, S.P.; Cámara, M.; Williams, P.; et al. Inhibition and Dispersion of Pseudomonas aeruginosa Biofilms by Glycopeptide Dendrimers Targeting the Fucose-Specific Lectin LecB. Chem. Biol. 2008, 15, 1249–1257. [Google Scholar] [CrossRef]
- Krachler, A.M.; Orth, K. Targeting the bacteria–host interface. Virulence 2013, 4, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Boas, D.P.A.V.; Sillankorva, S.; Azeredo, J. Phage Therapy: A Step Forward in the Treatment of Pseudomonas aeruginosa Infections. J. Virol. 2015, 89, 7449–7456. [Google Scholar] [CrossRef] [Green Version]
- Vandenheuvel, D.; Lavigne, R.; Brüssow, H. Bacteriophage Therapy: Advances in Formulation Strategies and Human Clinical Trials. Annu. Rev. Virol. 2015, 2, 599–618. [Google Scholar] [CrossRef]
- Penadés, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef]
- Hesse, S.E.; Adhya, S. Phage Therapy in the Twenty-First Century: Facing the Decline of the Antibiotic Era; Is It Finally Time for the Age of the Phage? Annu. Rev. Microbiol. 2019, 73, 155–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melander, R.J.; Basak, A.K.; Melander, C. Natural products as inspiration for the development of bacterial antibiofilm agents. Nat. Prod. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Salomoni, R.; Léo, P.; Montemor, A.; Rinaldi, B.; Rodrigues, M. Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa. Nanotechnol. Sci. Appl. 2017, 10, 115–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Shah, K.N.; Zhang, F.; Salazar, A.J.; Shah, P.N.; Li, R.; Sacchettini, J.; Wooley, K.L.; Cannon, C.L. Minocycline and Silver Dual-Loaded Polyphosphoester-Based Nanoparticles for Treatment of Resistant Pseudomonas aeruginosa. Mol. Pharm. 2019, 16, 1606–1619. [Google Scholar] [CrossRef] [PubMed]
- Türeli, N.G.; Torge, A.; Juntke, J.; Schwarz, B.C.; Schneider-Daum, N.; Türeli, A.E.; Lehr, C.-M.; Schneider, M. Ciprofloxacin-loaded PLGA nanoparticles against cystic fibrosis P. aeruginosa lung infections. Eur. J. Pharm. Biopharm. 2017, 117, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Deacon, J.; Abdelghany, S.M.; Quinn, D.J.; Schmid, D.; Megaw, J.; Donnelly, R.F.; Jones, D.S.; Kissenpfennig, A.; Elborn, J.S.; Gilmore, B.F.; et al. Antimicrobial efficacy of tobramycin polymeric nanoparticles for Pseudomonas aeruginosa infections in cystic fibrosis: Formulation, characterisation and functionalisation with dornase alfa (DNase). J. Control. Release 2015, 198, 55–61. [Google Scholar] [CrossRef] [Green Version]
- D’Angelo, I.; Casciaro, B.; Miro, A.; Quaglia, F.; Mangoni, M.L.; Ungaro, F. Overcoming barriers in Pseudomonas aeruginosa lung infections: Engineered nanoparticles for local delivery of a cationic antimicrobial peptide. Colloids Surf. B Biointerfaces 2015, 135, 717–725. [Google Scholar] [CrossRef]
- Baelo, A.; Levato, R.; Julián, E.; Crespo, A.; Astola, J.; Gavaldà, J.; Engel, E.; Mateos-Timoneda, M.A.; Torrents, E. Disassembling bacterial extracellular matrix with DNase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J. Control. Release 2015, 209, 150–158. [Google Scholar] [CrossRef]
- Powell, L.C.; Pritchard, M.F.; Ferguson, E.L.; Powell, K.A.; Patel, S.U.; Rye, P.D.; Sakellakou, S.-M.; Buurma, N.J.; Brilliant, C.D.; Copping, J.M.; et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes 2018, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Oh, H.-S.; Ng, Y.C.B.; Tang, P.Y.P.; Barraud, N.; Rice, S.A. Nitric Oxide-Mediated Induction of Dispersal in Pseudomonas aeruginosa Biofilms Is Inhibited by Flavohemoglobin Production and Is Enhanced by Imidazole. Antimicrob. Agents Chemother. 2017, 62, e01832-17. [Google Scholar] [CrossRef] [Green Version]
- Birmes, F.S.; Säring, R.; Hauke, M.C.; Ritzmann, N.H.; Drees, S.L.; Daniel, J.; Treffon, J.; Liebau, E.; Kahl, B.C.; Fetzner, S. Interference with Pseudomonas aeruginosa Quorum Sensing and Virulence by the Mycobacterial Pseudomonas Quinolone Signal Dioxygenase AqdC in Combination with the N-Acylhomoserine Lactone Lactonase QsdA. Infect. Immun. 2019, 87, e00278-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Park, J.-S.; Choi, H.-Y.; Yoon, S.S.; Kim, W.-G. Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: a connection between quorum sensing and c-di-GMP. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kiymaci, M.E.; Altanlar, N.; Gumustas, M.; Ozkan, S.A.; Akin, A. Quorum sensing signals and related virulence inhibition of Pseudomonas aeruginosa by a potential probiotic strain’s organic acid. Microb. Pathog. 2018, 121, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, Y.; Thoendel, M.; Olakanmi, O.; Britigan, B.E.; Singh, P.K. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Investig. 2007, 117, 877–888. [Google Scholar] [CrossRef]
- Moreau-Marquis, S.; O’Toole, G.A.; Stanton, B.A. Tobramycin and FDA-Approved Iron Chelators Eliminate Pseudomonas aeruginosa Biofilms on Cystic Fibrosis Cells. Am. J. Respir. Cell Mol. Biol. 2009, 41, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Soukos, N.S.; Goodson, J.M. Photodynamic therapy in the control of oral biofilms. Periodontol 2000 2010, 55, 143–166. [Google Scholar] [CrossRef]
- Kübler, A.C. Photodynamic therapy. Med Laser Appl. 2005, 20, 37–45. [Google Scholar] [CrossRef]
- Orlandi, V.T.; Rybtke, M.; Caruso, E.; Banfi, S.; Tolker-Nielsen, T.; Barbieri, P. Antimicrobial and anti-biofilm effect of a novel BODIPY photosensitizer against Pseudomonas aeruginosa PAO1. Biofouling 2014, 30, 883–891. [Google Scholar] [CrossRef]
- Vassena, C.; Fenu, S.; Giuliani, F.; Fantetti, L.; Roncucci, G.; Simonutti, G.; Romanò, C.L.; De Francesco, R.; Drago, L. Photodynamic antibacterial and antibiofilm activity of RLP068/Cl against Staphylococcus aureus and Pseudomonas aeruginosa forming biofilms on prosthetic material. Int. J. Antimicrob. Agents 2014, 44, 47–55. [Google Scholar] [CrossRef]
- Parasuraman, P.; Antony, A.P.; Sruthil, L.S.B.; Sharan, A.; Siddhardha, B.; Kasinathan, K.; Bahkali, N.A.; Dawoud, T.M.S.; Syed, A. Antimicrobial photodynamic activity of toluidine blue encapsulated in mesoporous silica nanoparticles against Pseudomonas aeruginosa and Staphylococcus aureus. Biofouling 2019, 35, 89–103. [Google Scholar] [CrossRef] [Green Version]
- Al-Bakri, A.G.; Mahmoud, N.N. Photothermal-Induced Antibacterial Activity of Gold Nanorods Loaded into Polymeric Hydrogel against Pseudomonas aeruginosa Biofilm. Molecules 2019, 24, 2661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Borg, R.E.; Dow, L.P.; Pruitt, B.L.; Chen, I.A. Controlled phage therapy by photothermal ablation of specific bacterial species using gold nanorods targeted by chimeric phages. Proc. Natl. Acad. Sci. USA 2020, 117, 1951–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Wang, H.; Dong, W.; Cheng, S.; Li, H.; Tan, J.; Zhou, J.; He, W.; Li, L.; Zhang, J.; et al. A multifunctional platform with single-NIR-laser-triggered photothermal and NO release for synergistic therapy against multidrug-resistant Gram-negative bacteria and their biofilms. J. Nanobiotechnol. 2020, 18, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Bilici, K.; Atac, N.; Muti, A.; Baylam, I.; Dogan, O.; Sennaroglu, A.; Can, F.; Acar, H.Y. Broad spectrum antibacterial photodynamic and photothermal therapy achieved with indocyanine green loaded SPIONs under near infrared irradiation. Biomater. Sci. 2020, 8, 4616–4625. [Google Scholar] [CrossRef]
- Wong, J.P.; Yang, H.; Blasetti, K.L.; Schnell, G.; Conley, J.; Schofield, L.N. Liposome delivery of ciprofloxacin against intracellular Francisella tularensis infection. J. Control Release 2003, 92, 265–273. [Google Scholar] [CrossRef]
- Varshosaz, J.; Ghaffari, S.; Saadat, A.; Atyabi, F. Stability and antimicrobial effect of amikacin-loaded solid lipid nanoparticles. Int. J. Nanomed. 2010, 6, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargonia, A.; Cavallib, R.; Zara, G.P.; Fundaròc, A.; Caputob, O.; Gasco, M.R. Transmucosal transport of tobramycin incorporated in solid lipid nanoparticles (sln) after duodenal administration to rats. Part II—Tissue distribution. Pharmacol. Res. 2001, 43, 497–502. [Google Scholar] [CrossRef]
- Alizadeh, A.; Salouti, M.; Alizadeh, H.; Kazemizadeh, A.R.; Safari, A.A.; Mahmazi, S. Enhanced antibacterial effect of azlocillin in conjugation with silver nanoparticles against Pseudomonas aeruginosa. IET Nanobiotechnol. 2017, 11, 942–947. [Google Scholar] [CrossRef]
- Lamppa, J.W.; Griswold, K.E. Alginate Lyase Exhibits Catalysis-Independent Biofilm Dispersion and Antibiotic Synergy. Antimicrob. Agents Chemother. 2012, 57, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Pleszczyńska, M.; Wiater, A.; Janczarek, M.; Szczodrak, J. (1→3)-α-d-Glucan hydrolases in dental biofilm prevention and control: A review. Int. J. Biol. Macromol. 2015, 79, 761–778. [Google Scholar] [CrossRef]
- Eckhart, L.; Fischer, H.; Barken, K.B.; Tolker-Nielsen, T.; Tschachler, E. DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Br. J. Dermatol. 2007, 156, 1342–1345. [Google Scholar] [CrossRef] [PubMed]
- Okshevsky, M.; Regina, V.R.; Meyer, R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kamesh, A.C.; Xiao, Y.; Sun, V.; Hayes, M.; Daniell, H.; Koo, H. Topical delivery of low-cost protein drug candidates made in chloroplasts for biofilm disruption and uptake by oral epithelial cells. Biomaterials 2016, 105, 156–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, E.; Seminara, A.B.; Kim, S.-K.; Hall, C.L.; Wang, Y.; Lee, V.T. Thiol-benzo-triazolo-quinazolinone Inhibits Alg44 Binding to c-di-GMP and Reduces Alginate Production by Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 3076–3085. [Google Scholar] [CrossRef]
- Warrener, P.; Varkey, R.; Bonnell, J.C.; DiGiandomenico, A.; Camara, M.; Cook, K.; Peng, L.; Zha, J.; Chowdury, P.; Sellman, B.; et al. A Novel Anti-PcrV Antibody Providing Enhanced Protection against Pseudomonas aeruginosa in Multiple Animal Infection Models. Antimicrob. Agents Chemother. 2014, 58, 4384–4391. [Google Scholar] [CrossRef] [Green Version]
- DiGiandomenico, A.; Warrener, P.; Hamilton, M.; Guillard, S.; Ravn, P.; Minter, R.; Camara, M.M.; Venkatraman, V.; MacGill, R.S.; Lin, J.; et al. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J. Exp. Med. 2012, 209, 1273–1287. [Google Scholar] [CrossRef] [Green Version]
- Novotny, L.A.; Jurcisek, J.A.; Goodman, S.D.; Bakaletz, L.O. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine 2016, 10, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Howlin, R.P.; Cathie, K.; Hall-Stoodley, L.; Cornelius, V.; Duignan, C.; Allan, R.N.; Fernandez, B.O.; Barraud, N.; Bruce, K.D.; Jefferies, J.; et al. Low-Dose Nitric Oxide as Targeted Anti-biofilm Adjunctive Therapy to Treat Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. Mol. Ther. 2017, 25, 2104–2116. [Google Scholar] [CrossRef] [Green Version]
- Chua, S.L.; Liu, Y.; Li, Y.; Ting, H.J.; Kohli, G.S.; Cai, Z.; Suwanchaikasem, P.; Goh, K.K.K.; Ng, S.P.; Tolker-Nielsen, T.; et al. Reduced Intracellular c-di-GMP Content Increases Expression of Quorum Sensing-Regulated Genes in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017, 7, 451. [Google Scholar] [CrossRef]
- Ouyang, J.; Sun, F.; Feng, W.; Sun, Y.; Qiu, X.; Xiong, L.; Liu, Y.; Chen, Y. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J. Appl. Microbiol. 2016, 120, 966–974. [Google Scholar] [CrossRef] [Green Version]
- Maura, D.; Rahme, L.G. Pharmacological Inhibition of the Pseudomonas aeruginosa MvfR Quorum-Sensing System Interferes with Biofilm Formation and Potentiates Antibiotic-Mediated Biofilm Disruption. Antimicrob. Agents Chemother. 2017, 61, e01362-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utari, P.D.; Setroikromo, R.; Melgert, B.N.; Quax, W.J. PvdQ Quorum Quenching Acylase Attenuates Pseudomonas aeruginosa Virulence in a Mouse Model of Pulmonary Infection. Front. Cell. Infect. Microbiol. 2018, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Oglesby-Sherrouse, A.G.; Djapgne, L.; Nguyen, A.T.; Vasil, A.I.; Vasil, M.L. The complex interplay of iron, biofilm formation, and mucoidy affecting antimicrobial resistance of Pseudomonas aeruginosa. Pathog. Dis. 2014, 70, 307–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Terwilliger, A.; Maresso, A.W. Iron and zinc exploitation during bacterial pathogenesis. Metallomics 2015, 7, 1541–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, D.W.; Carroll, V.; O’May, C.Y.; Champion, A.; Kirov, S.M. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur. Respir. J. 2007, 30, 286–292. [Google Scholar] [CrossRef]
- Banin, E.; Lozinski, A.; Brady, K.M.; Berenshtein, E.; Butterfield, P.W.; Moshe, M.; Chevion, M.; Greenberg, E.P. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. USA 2008, 105, 16761–16766. [Google Scholar] [CrossRef] [Green Version]
- Darabpour, E.; Kashef, N.; Mashayekhan, S. Chitosan nanoparticles enhance the efficiency of methylene blue-mediated antimicrobial photodynamic inactivation of bacterial biofilms: An in vitro study. Photodiagnosis Photodyn. Ther. 2016, 14, 211–217. [Google Scholar] [CrossRef]
- Wainwright, M.; Phoenix, D.; Laycock, S.; Wareing, D.; Wright, P. Photobactericidal activity of phenothiazinium dyes against methicillin-resistant strains of Staphylococcus aureus. FEMS Microbiol. Lett. 1998, 160, 177–181. [Google Scholar] [CrossRef]
- Vatansever, F.; De Melo, W.C.; Avci, P.; Vecchio, D.; Sadasivam, M.; Gupta, A.; Chandran, R.; Karimi, M.; Parizotto, N.A.; Yin, R.; et al. Antimicrobial strategies centered around reactive oxygen species—Bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 2013, 37, 955–989. [Google Scholar] [CrossRef] [Green Version]
- Tavares, A.; Carvalho, C.M.B.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Tome, J.P.C.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Gomes, N.C.; Alves, E.; et al. Antimicrobial Photodynamic Therapy: Study of Bacterial Recovery Viability and Potential Development of Resistance after Treatment. Mar. Drugs 2010, 8, 91–105. [Google Scholar] [CrossRef] [Green Version]
- Misba, L.; Zaidi, S.; Khan, A.U. A comparison of antibacterial and antibiofilm efficacy of phenothiazinium dyes between Gram positive and Gram negative bacterial biofilm. Photodiagnosis Photodyn. Ther. 2017, 18, 24–33. [Google Scholar] [CrossRef]
- Beirão, S.; Fernandes, S.; Coelho, J.; Faustino, M.A.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Tomé, A.C.; Almeida, A. Cunha, Ângela Photodynamic Inactivation of Bacterial and Yeast Biofilms with a Cationic Porphyrin. Photochem. Photobiol. 2014, 90, 1387–1396. [Google Scholar] [CrossRef]
- Zhao, Y.; Lu, Z.; Dai, X.; Wei, X.; Yu, Y.; Chen, X.; Zhang, X.; Li, C. Glycomimetic-Conjugated Photosensitizer for Specific Pseudomonas aeruginosa Recognition and Targeted Photodynamic Therapy. Bioconjugate Chem. 2018, 29, 3222–3230. [Google Scholar] [CrossRef]
- Abdulrahman, H.; Misba, L.; Ahmad, S.; Khan, A.U. Curcumin induced photodynamic therapy mediated suppression of quorum sensing pathway of Pseudomonas aeruginosa: An approach to inhibit biofilm in vitro. Photodiagnosis Photodyn. Ther. 2020, 30, 101645. [Google Scholar] [CrossRef]
- Korupalli, C.; Huang, C.-C.; Lin, W.-C.; Pan, W.-Y.; Lin, P.-Y.; Wan, W.-L.; Li, M.-J.; Chang, Y.; Sung, H. Acidity-triggered charge-convertible nanoparticles that can cause bacterium-specific aggregation in situ to enhance photothermal ablation of focal infection. Biomaterials 2017, 116, 1–9. [Google Scholar] [CrossRef]
- Murali, V.S.; Wang, R.; Mikoryak, C.A.; Pantano, P.; Draper, R. The impact of subcellular location on the near infrared-mediated thermal ablation of cells by targeted carbon nanotubes. Nanotechnology 2016, 27, 425102. [Google Scholar] [CrossRef] [Green Version]
- Qian, W.; Yan, C.; He, D.; Yu, X.; Yuan, L.; Liu, M.; Wang, S.; Deng, J. pH-triggered charge-reversible of glycol chitosan conjugated carboxyl graphene for enhancing photothermal ablation of focal infection. Acta Biomater. 2018, 69, 256–264. [Google Scholar] [CrossRef]
- Behzadpour, N.; Sattarahmady, N.; Akbari, N. Antimicrobial Photothermal Treatment of Pseudomonas Aeruginosa by a Carbon Nanoparticles-Polypyrrole Nanocomposite. J. Biomed. Phys. Eng. 2019, 9, 661–672. [Google Scholar] [CrossRef]
- Dickerson, E.B.; Dreaden, E.C.; Huang, X.; El-Sayed, I.H.; Chu, H.; Pushpanketh, S.; McDonald, J.F.; El-Sayed, M.A. Gold nanorod assisted near-infrared plasmonic photothermal therapy (PPTT) of squamous cell carcinoma in mice. Cancer Lett. 2008, 269, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Zhou, J.; Zhuang, J.; Gao, H.; Huang, D.; Wang, L.; Wu, W.; Li, Q.; Yang, D.-P.; Han, M.-Y. Strong Near-Infrared Absorbing and Biocompatible CuS Nanoparticles for Rapid and Efficient Photothermal Ablation of Gram-Positive and -Negative Bacteria. ACS Appl. Mater. Interfaces 2017, 9, 36606–36614. [Google Scholar] [CrossRef]
- Abel, S.B.; A Gallarato, L.; Dardanelli, M.S.; A Barbero, C.; Rivarola, C.; Yslas, E.I. Photothermal lysis of Pseudomonas aeruginosa by polyaniline nanoparticles under near infrared irradiation. Biomed. Phys. Eng. Express 2018, 4, 045037. [Google Scholar] [CrossRef] [Green Version]


| Therapeutic Approach | Activity | Advantages | Limitation | References |
|---|---|---|---|---|
| Antimicrobial peptides |
|
|
| [139,140,141,142,143,144] |
| Antibiotics |
|
|
| [145] |
| Lectin inhibitor |
|
|
| [146,147,148] |
| Bacteriophages |
|
|
| [149,150,151,152,153] |
| Natural products |
|
|
| [154] |
| Nanoparticles |
|
|
| [155,156] |
| Nanocarriers (Liposomes, solid lipid and polymeric) |
|
|
| [157,158,159] |
| EPS inhibitors |
|
|
| [35,160,161] |
| Biofilm dispersers |
|
|
| [66,162] |
| QS inhibitors |
|
|
| [163,164,165] |
| Iron chelator |
|
|
| [166,167] |
| Photodynamic therapy |
|
|
| [168,169,170,171,172] |
| Photothermal therapy |
|
|
| [173,174,175,176] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. https://doi.org/10.3390/ijms21228671
Thi MTT, Wibowo D, Rehm BHA. Pseudomonas aeruginosa Biofilms. International Journal of Molecular Sciences. 2020; 21(22):8671. https://doi.org/10.3390/ijms21228671
Chicago/Turabian StyleThi, Minh Tam Tran, David Wibowo, and Bernd H.A. Rehm. 2020. "Pseudomonas aeruginosa Biofilms" International Journal of Molecular Sciences 21, no. 22: 8671. https://doi.org/10.3390/ijms21228671
APA StyleThi, M. T. T., Wibowo, D., & Rehm, B. H. A. (2020). Pseudomonas aeruginosa Biofilms. International Journal of Molecular Sciences, 21(22), 8671. https://doi.org/10.3390/ijms21228671