Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes
Abstract
:1. Introduction
2. Results
2.1. Neutralization of Nucleic Acids Is Essential for Their Efficient Transfer by Fusogenic Liposomes
2.2. Effect of the Liposomal Formulation on Nucleic Acid Transfer Efficiency
2.3. Quantification of mRNA Transfer on Single Cell Level
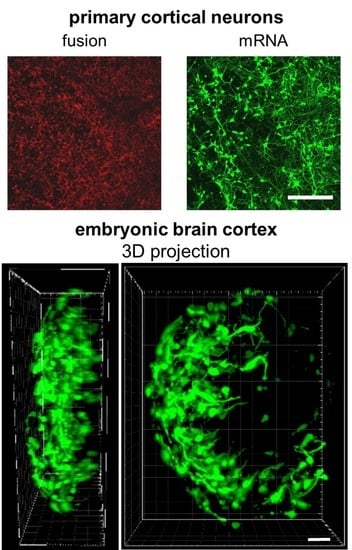
2.4. mRNA Delivery Into Primary Cortical Neurons and Freshly Isolated Neural Tissue
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Liposomes
4.3. Preparation of Neutralization Reagents (NR)
4.4. Preparation of Nucleic Acid Containing Fusogenic Liposomes
4.5. Quantification of Knock Down Efficiency of Transiently Expressed eGFP
4.6. mRNA Transfer Into Isolated Cortical Brain Tissue
4.7. Zeta Potential and Size Distribution Measurements
4.8. Light Microscopy
4.9. Flow Cytometry
4.10. Quantitative Analysis of Membrane Fusion Events
4.11. RNA Isolation and cDNA Synthesis
4.12. RT-PCR Assays
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.M.; Erdman, Z.S.; Kokkoli, E. Transfection mechanisms of polyplexes, lipoplexes, and stealth liposomes in alpha(5)beta(1) integrin bearing DLD-1 colorectal cancer cells. Langmuir 2014, 30, 3802–3810. [Google Scholar] [CrossRef]
- Kaczmarek, J.C.; Patel, A.K.; Kauffman, K.J.; Fenton, O.S.; Webber, M.J.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew Chem. Int. Ed. Engl. 2016, 55, 13808–13812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinnapireddy, S.R.; Duse, L.; Strehlow, B.; Schafer, J.; Bakowsky, U. Composite liposome-PEI/nucleic acid lipopolyplexes for safe and efficient gene delivery and gene knockdown. Colloids Surf. B Biointerfaces 2017, 158, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Wolk, C.; Janich, C.; Meister, A.; Drescher, S.; Langner, A.; Brezesinski, G.; Bakowsky, U. Investigation of Binary Lipid Mixtures of a Three-Chain Cationic Lipid with Phospholipids Suitable for Gene Delivery. Bioconjug. Chem. 2015, 26, 2461–2473. [Google Scholar] [CrossRef] [PubMed]
- Kafri, T. Gene delivery by lentivirus vectors an overview. Methods Mol. Biol. 2004, 246, 367–390. [Google Scholar]
- Cockrell, A.S.; Kafri, T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007, 36, 184–204. [Google Scholar] [CrossRef]
- Zuhorn, I.S.; Kalicharan, R.; Hoekstra, D. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J. Biol. Chem. 2002, 277, 18021–18028. [Google Scholar] [CrossRef] [Green Version]
- Rappoport, J.Z. Focusing on clathrin-mediated endocytosis. Biochem. J. 2008, 412, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Cardarelli, F.; Digiacomo, L.; Marchini, C.; Amici, A.; Salomone, F.; Fiume, G.; Rossetta, A.; Gratton, E.; Pozzi, D.; Caracciolo, G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016, 6, 25879. [Google Scholar] [CrossRef] [Green Version]
- Selby, L.I.; Cortez-Jugo, C.M.; Such, G.K.; Johnston, A.P.R. Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Lonn, P.; Kacsinta, A.D.; Cui, X.S.; Hamil, A.S.; Kaulich, M.; Gogoi, K.; Dowdy, S.F. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep. 2016, 6, 32301. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamada, K.; Fujiwara, Y.; Sato, Y.; Harashima, H. Reducing the Cytotoxicity of Lipid Nanoparticles Associated with a Fusogenic Cationic Lipid in a Natural Killer Cell Line by Introducing a Polycation-Based siRNA Core. Mol. Pharm. 2018, 15, 2142–2150. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorgi, F.L.; Bhattacharya, S.; Huang, L. Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther. 1997, 4, 961–968. [Google Scholar] [CrossRef]
- Skold, A.E.; van Beek, J.J.; Sittig, S.P.; Bakdash, G.; Tel, J.; Schreibelt, G.; de Vries, I.J. Protamine-stabilized RNA as an ex vivo stimulant of primary human dendritic cell subsets. Cancer Immunol. Immunother. 2015, 64, 1461–1473. [Google Scholar] [CrossRef] [Green Version]
- Mao, H.Q.; Roy, K.; Troung-Le, V.L.; Janes, K.A.; Lin, K.Y.; Wang, Y.; August, J.T.; Leong, K.W. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J. Control. Release 2001, 70, 399–421. [Google Scholar] [CrossRef]
- Tezgel, O.; Szarpak-Jankowska, A.; Arnould, A.; Auzely-Velty, R.; Texier, I. Chitosan-lipid nanoparticles (CS-LNPs): Application to siRNA delivery. J. Colloid Interface Sci. 2018, 510, 45–56. [Google Scholar] [CrossRef]
- Mukalel, A.J.; Riley, R.S.; Zhang, R.; Mitchell, M.J. Nanoparticles for nucleic acid delivery: Applications in cancer immunotherapy. Cancer Lett. 2019, 458, 102–112. [Google Scholar] [CrossRef]
- Lee, K.D.; Nir, S.; Papahadjopoulos, D. Quantitative analysis of liposome-cell interactions in vitro: rate constants of binding and endocytosis with suspension and adherent J774 cells and human monocytes. Biochemistry 1993, 32, 889–899. [Google Scholar] [CrossRef]
- Hama, S.; Akita, H.; Ito, R.; Mizuguchi, H.; Hayakawa, T.; Harashima, H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol.Ther. 2006, 13, 786–794. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Atobe, K.; Barichello, J.M.; Ishida, T.; Kiwada, H. Complex formation with plasmid DNA increases the cytotoxicity of cationic liposomes. Biol. Pharm. Bull. 2007, 30, 751–757. [Google Scholar] [CrossRef] [Green Version]
- Lonez, C.; Vandenbranden, M.; Elouahabi, A.; Ruysschaert, J.M. Cationic lipid/DNA complexes induce TNF-alpha secretion in splenic macrophages. Eur. J. Pharm. Biopharm. 2008, 69, 817–823. [Google Scholar] [CrossRef]
- Xia, T.; Kovochich, M.; Liong, M.; Zink, J.I.; Nel, A.E. Cationic polystyrene nanosphere toxicity depends on cell-specific endocytic and mitochondrial injury pathways. ACS Nano 2008, 2, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brencicova, E.; Diebold, S.S. Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Front. Cell Infect. Microbiol. 2013, 3, 37. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef] [Green Version]
- Elegheert, J.; Behiels, E.; Bishop, B.; Scott, S.; Woolley, R.E.; Griffiths, S.C.; Byrne, E.F.X.; Chang, V.T.; Stuart, D.I.; Jones, E.Y.; et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat. Protoc. 2018, 13, 2991–3017. [Google Scholar] [CrossRef]
- Ellis, B.L.; Hirsch, M.L.; Barker, J.C.; Connelly, J.P.; Steininger, R.J., 3rd; Porteus, M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013, 10, 74. [Google Scholar] [CrossRef] [Green Version]
- Csiszar, A.; Hersch, N.; Dieluweit, S.; Biehl, R.; Merkel, R.; Hoffmann, B. Novel fusogenic liposomes for fluorescent cell labeling and membrane modification. Bioconjug. Chem. 2010, 21, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Kleusch, C.; Hersch, N.; Hoffmann, B.; Merkel, R.; Csiszar, A. Fluorescent lipids: functional parts of fusogenic liposomes and tools for cell membrane labeling and visualization. Molecules 2012, 17, 1055–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, T.; Kleusch, C.; Naumovska, E.; Merkel, R.; Csiszar, A. A bioanalytical assay to distinguish cellular uptake routes for liposomes. Cytom. A 2016, 89, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Kube, S.; Hersch, N.; Naumovska, E.; Gensch, T.; Hendriks, J.; Franzen, A.; Landvogt, L.; Siebrasse, J.P.; Kubitscheck, U.; Hoffmann, B.; et al. Fusogenic Liposomes as Nanocarriers for the Delivery of Intracellular Proteins. Langmuir 2017, 33, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Ramms, L.; Fabris, G.; Windoffer, R.; Schwarz, N.; Springer, R.; Zhou, C.; Lazar, J.; Stiefel, S.; Hersch, N.; Schnakenberg, U.; et al. Keratins as the main component for the mechanical integrity of keratinocytes. Proc. Natl. Acad. Sci. USA 2013, 110, 18513–18518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumovska, E.; Ludwanowski, S.; Hersch, N.; Braun, T.; Merkel, R.; Hoffmann, B.; Csiszar, A. Plasma membrane functionalization using highly fusogenic immune activator liposomes. Acta Biomater. 2014, 10, 1403–1411. [Google Scholar] [CrossRef]
- Hoffmann, M.e.a. Changing the Way of Entrance: Highly Efficient Transfer of mRNA and siRNA via Fusogenic Nano-Carriers. J. Biomed. Nanotechnol. 2019, 15, 14. [Google Scholar] [CrossRef]
- Hersch, N.; Wolters, B.; Ungvari, Z.; Gautam, T.; Deshpande, D.; Merkel, R.; Csiszar, A.; Hoffmann, B.; Csiszar, A. Biotin-conjugated fusogenic liposomes for high-quality cell purification. J. Biomater. Appl. 2016, 30, 846–856. [Google Scholar] [CrossRef]
- Mayhew, E.; Juliano, R. Interaction of polynucleotides with cultured mammalian cells. II. Cell surface charge density and RNA uptake. Exp. Cell Res. 1973, 77, 409–414. [Google Scholar] [CrossRef]
- Regelin, A.E.; Fankhaenel, S.; Gurtesch, L.; Prinz, C.; von Kiedrowski, G.; Massing, U. Biophysical and lipofection studies of DOTAP analogs. Biochim. Biophys. Acta 2000, 1464, 151–164. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, L. DNA transfection mediated by cationic liposomes containing lipopolylysine: characterization and mechanism of action. Biochim. Biophys. Acta 1994, 1189, 195–203. [Google Scholar] [CrossRef]
- Kolasinac, R.; Kleusch, C.; Braun, T.; Merkel, R.; Csiszar, A. Deciphering the Functional Composition of Fusogenic Liposomes. Int. J. Mol. Sci. 2018, 19, 346. [Google Scholar] [CrossRef] [Green Version]
- Gresch, O.; Altrogge, L. Transfection of difficult-to-transfect primary mammalian cells. Methods Mol. Biol. 2012, 801, 65–74. [Google Scholar] [CrossRef]
- Zhang, H.S.; Kim, E.; Lee, S.; Ahn, I.S.; Jang, J.H. Transduction of striatum and cortex tissues by adeno-associated viral vectors produced by herpes simplex virus- and baculovirus-based methods. J. Virol. Methods 2012, 179, 276–280. [Google Scholar] [CrossRef]
- Ewert, K.K.; Zidovska, A.; Ahmad, A.; Bouxsein, N.F.; Evans, H.M.; McAllister, C.S.; Samuel, C.E.; Safinya, C.R. Cationic liposome-nucleic acid complexes for gene delivery and silencing: pathways and mechanisms for plasmid DNA and siRNA. Top. Curr. Chem. 2010, 296, 191–226. [Google Scholar]
- David, S.; Passirani, C.; Carmoy, N.; Morille, M.; Mevel, M.; Chatin, B.; Benoit, J.P.; Montier, T.; Pitard, B. DNA nanocarriers for systemic administration: characterization and in vivo bioimaging in healthy mice. Mol. Nucleic Acids 2013, 2, e64. [Google Scholar] [CrossRef]
- Safinya, C.R.; Ewert, K.K.; Majzoub, R.N.; Leal, C. Cationic liposome-nucleic acid complexes for gene delivery and gene silencing. New J. Chem. 2014, 38, 5164–5172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munroe, D.; Jacobson, A. mRNA poly(A) tail, a 3’ enhancer of translational initiation. Mol. Cell Biol. 1990, 10, 3441–3455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicol, F.; Nir, S.; Szoka, F.C., Jr. Effect of cholesterol and charge on pore formation in bilayer vesicles by a pH-sensitive peptide. Biophys. J. 1996, 71, 3288–3301. [Google Scholar] [CrossRef] [Green Version]
- Nicol, F.; Nir, S.; Szoka, F.C., Jr. Effect of phospholipid composition on an amphipathic peptide-mediated pore formation in bilayer vesicles. Biophys. J. 2000, 78, 818–829. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.Q.; Chen, J.L.; Lv, T.F.; He, C.X.; Tang, G.P.; Liang, W.Q.; Tabata, Y.; Gao, J.Q. N/P ratio significantly influences the transfection efficiency and cytotoxicity of a polyethylenimine/chitosan/DNA complex. Biol. Pharm. Bull. 2009, 32, 706–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Zeng, X.; Liu, M.; Deng, Y.; He, N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics 2014, 4, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolasinac, R.; Jaksch, S.; Dreissen, G.; Braeutigam, A.; Merkel, R.; Csiszar, A. Influence of Environmental Conditions on the Fusion of Cationic Liposomes with Living Mammalian Cells. Nanomaterials 2019, 9, 1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rettig, L.; Haen, S.P.; Bittermann, A.G.; von Boehmer, L.; Curioni, A.; Kramer, S.D.; Knuth, A.; Pascolo, S. Particle size and activation threshold: a new dimension of danger signaling. Blood 2010, 115, 4533–4541. [Google Scholar] [CrossRef] [Green Version]
- Kozielski, K.L.; Tzeng, S.Y.; De Mendoza, B.A.; Green, J.J. Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS Nano 2014, 8, 3232–3241. [Google Scholar] [CrossRef]
- Hagerman, P.J. Flexibility of RNA. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 139–156. [Google Scholar] [CrossRef]
- Son, K.K.; Tkach, D.; Patel, D.H. Zeta potential of transfection complexes formed in serum-free medium can predict in vitro gene transfer efficiency of transfection reagent. Biochim. Biophys. Acta 2000, 1468, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Lee, S.J.; Kim, J.K.; Choi, H.G.; Lim, S.J. Optimization and physicochemical characterization of a cationic lipid-phosphatidylcholine mixed emulsion formulated as a highly efficient vehicle that facilitates adenoviral gene transfer. Int. J. Nanomed. 2017, 12, 7323–7335. [Google Scholar] [CrossRef] [Green Version]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razinkov, V.I.; Melikyan, G.B.; Cohen, F.S. Hemifusion between cells expressing hemagglutinin of influenza virus and planar membranes can precede the formation of fusion pores that subsequently fully enlarge. Biophys. J. 1999, 77, 3144–3151. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Wang, J.; Ping, Q.; Yeo, Y. Quantitative Assessment of Nanoparticle Biodistribution by Fluorescence Imaging, Revisited. ACS Nano 2018, 12, 6458–6468. [Google Scholar] [CrossRef] [PubMed]
- Batard, P.; Jordan, M.; Wurm, F. Transfer of high copy number plasmid into mammalian cells by calcium phosphate transfection. Gene 2001, 270, 61–68. [Google Scholar] [CrossRef]
- Leonhardt, C.; Schwake, G.; Stogbauer, T.R.; Rappl, S.; Kuhr, J.T.; Ligon, T.S.; Radler, J.O. Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine 2014, 10, 679–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, J.A.; Linnartz, C.; Dreissen, G.; Springer, R.; Blaschke, S.; Rueger, M.A.; Fink, G.R.; Hoffmann, B.; Merkel, R. Directing Neuronal Outgrowth and Network Formation of Rat Cortical Neurons by Cyclic Substrate Stretch. Langmuir 2018. [Google Scholar] [CrossRef]






| NA | NA Length (Nucleotides) | Molar Ratio of NR/NA | Nucleotides/NR |
|---|---|---|---|
| siRNA | 20–30 | 5/1 | 4–6/1 |
| mRNA | 1000 | 50/1 | 20/1 |
| pDNA | 4700 | 450/1 | 16/1 |
| NR | Size [kDa] | NR/NA [mol/mol] |
|---|---|---|
| PEI branched | 25 | 0.4/1 |
| chitosan | 100 | 0.8/1 |
| PEI linear | 10 | 10/1 |
| protamine | 3–6 | 50/1 |
| pH of NR Buffer | Zeta Potential ± s.d. (mV) | Size ± s.d. (nm) | eGFP Transfer Efficiency ± s.d. (%) | eGFP Fluorescence Intensity ± s.d. (counts) |
|---|---|---|---|---|
| 11 | 60.5 ± 0.3 | 141.5 ± 36 | 54 ± 7 | 475 ± 117 |
| 8 | 54.5 ± 3 | 344.8± 20 | 46 ± 3 | 230 ± 88 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffmann, M.; Hersch, N.; Gerlach, S.; Dreissen, G.; Springer, R.; Merkel, R.; Csiszár, A.; Hoffmann, B. Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes. Int. J. Mol. Sci. 2020, 21, 2244. https://doi.org/10.3390/ijms21062244
Hoffmann M, Hersch N, Gerlach S, Dreissen G, Springer R, Merkel R, Csiszár A, Hoffmann B. Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes. International Journal of Molecular Sciences. 2020; 21(6):2244. https://doi.org/10.3390/ijms21062244
Chicago/Turabian StyleHoffmann, Marco, Nils Hersch, Sven Gerlach, Georg Dreissen, Ronald Springer, Rudolf Merkel, Agnes Csiszár, and Bernd Hoffmann. 2020. "Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes" International Journal of Molecular Sciences 21, no. 6: 2244. https://doi.org/10.3390/ijms21062244
APA StyleHoffmann, M., Hersch, N., Gerlach, S., Dreissen, G., Springer, R., Merkel, R., Csiszár, A., & Hoffmann, B. (2020). Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes. International Journal of Molecular Sciences, 21(6), 2244. https://doi.org/10.3390/ijms21062244