The Ionophoric Activity of a Pro-Apoptotic VEGF165 Fragment on HUVEC Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Peptides: Hydrophobicity and Conformational Analysis
2.2. Peptide Permeability on a Membrane Model System
2.3. Interaction with Plasma Membrane in Living Cells
2.4. Effects of Peptides on HUVEC: Viability and Apoptosis Assays
2.5. Effect of Copper (II) Ion on VEGFQ79G and VEGFI83G
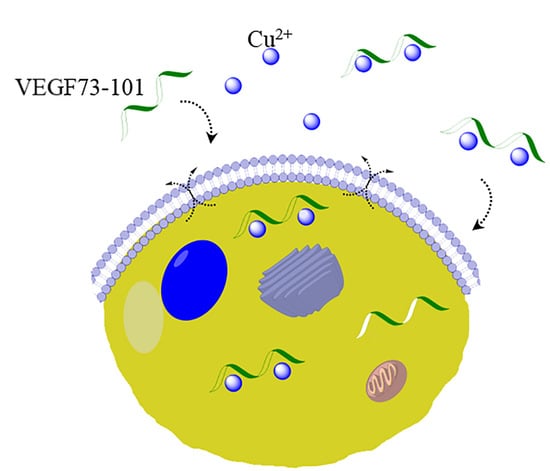
2.6. Model Membrane Assay of VEGF73-101/Cu(II) and Cellular Uptake
2.7. Effects of Peptide/Copper(II) Complexes on HUVEC: Viability and Apoptosis Assays
2.8. Effect of a Copper Chelator (BCS) on Cells HUVEC Treated with VEGF73-101/Cu(II)
3. Materials and Methods
3.1. Materials
3.2. Spectroscopic Measurements (CD e UV)
3.3. Cell Cultures
3.4. Cell Viability Assay (MTS)
3.5. Annexin V-FITC/PI Staining
3.6. Flow Cytometry
3.7. Interaction Peptides/Cell Membranes in Living Cells
3.8. Model Membrane Permeabilization Assays
3.9. Electrospray Mass Spectrometry (ESI-MS) Analysis
3.10. UHPLC-ESI MS
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Finney, L.; Vogt, S.; Fukai, T.; Glesne, D. Copper and angiogenesis: Unravelling a relationship key to cancer progression. Front. Res. 2009, 36, 88–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupte, A.; Mumper, R.J. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 2009, 35, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Cater, M.A.; Pearson, H.B.; Wolyniec, K.; Klaver, P.; Bilandzic, M.; Paterson, B.M.; Bush, A.I.; Humbert, P.O.; La Fontaine, S.; Donnelly, P.S.; et al. Increasing intracellular bioavailable copper selectively targets prostate cancer cells. Chem. Biol. 2013, 8, 1621–1631. [Google Scholar] [CrossRef]
- Kuo, H.W.; Chen, S.F.; Wu, C.C.; Chen, D.R.; Lee, J.H. Serum and tissue trace elements in patients with breast cancer in taiwan. Biol. Trace Element Res. 1999, 89, 1–11. [Google Scholar] [CrossRef]
- Tardito, S.; Marchiò, L. Copper compounds in anticancer strategies. Curr. Med. Chem. 2009, 16, 1325–1348. [Google Scholar] [CrossRef]
- Mahendiran, D.; Amuthakala, S.; Bhuvanesh, N.S.P.; Kumarc, R.S.; Rahiman, A.K. Copper complexes as prospective anticancer agents: In vitro and in vivo evaluation, selective targeting of cancer cells by DNA damage and Sphase arrest. RSC Adv. 2018, 8, 16973–16990. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.Y.; Wang, Y.N.; Liu, H.F.; Luo, Z.H.; Zhang, P.L.; Li-Fang, H.; Liu, M.R. Anti-cancer activities of metal-based complexes by regulating the VEGF/VEGFR2 signaling pathway and apoptosis-related factors Bcl-2, Bax, and caspase-9 to inhibit angiogenesis and induce apoptosis. Metallomics 2020, 12, 92–103. [Google Scholar] [CrossRef]
- Sen, C.K.; Khanna, S.; Venojarvi, M.; Trikha, P.; Ellison, E.C.; Hunt, T.K.; Roy, S. Copper-induced vascular endothelial growth factor expression and wound healing. Am. J. Physiol. Heart Circ. Physiol. 2002, 282, H1821–H1827. [Google Scholar] [CrossRef] [Green Version]
- Martin, F.; Linden, T.; Katschinski, D.M.; Oehme, F.; Flamme, I.; Mukhopadhyay, C.K.; Eckhardt, K.; Tröger, J.; Barth, S.; Camenisch, G.; et al. Copper-dependent activation of hypoxia-inducible factor (HIF)-1: Implications for ceruloplasmin regulation. Blood. 2005, 105, 4613–4619. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.; Bao, L.W.; Merajver, S.D. Tetrathiomolybdate inhibits angiogenesis and metastasis through suppression of the NFkappaB signaling cascade. Mol. Cancer Res. 2003, 1, 701–706. [Google Scholar]
- Feng, W.; Ye, F.; Xue, W.; Zhou, Z.; Kang, Y.J. Copper regulation of hypoxia-inducible factor-1 activity. Mol. Pharmacol. 2009, 75, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Jazayeri, S.; Feli, A.; Bitaraf, M.A.; Solaymani Dodaran, M.; Alikhani, M.; Hosseinzadeh-Attar, M.J. Effects of copper reduction on angiogenesis-related factors in recurrent glioblastoma cases. Asian Pac. J. Cancer Prev. 2016, 17, 4609–4614. [Google Scholar] [CrossRef]
- Frezza, M.; Hindo, S.; Chen, D.; Davenport, A.; Schmitt, S.; Tomco, D.; Dou, Q.P. Novel metals and metal complexes as platforms for cancer therapy. Curr. Pharm. Des. 2010, 16, 1813–1825. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.; Kleer, C.G.; van Golen, K.L.; Irani, J.; Bottema, K.M.; Bias, C.; De Carvalho, M.; Mesri, E.A.; Robins, D.M.; Dick, R.D.; et al. Copper deficiency induced by tetrathiomolybdate suppresses tumor growth and angiogenesis. Cancer Res. 2002, 62, 4854–4859. [Google Scholar]
- Lowndes, S.A.; Harris, A.L. The role of copper in tumour angiogenesis. J. Mammary Gland Biol. Neoplasia 2005, 10, 299–310. [Google Scholar] [CrossRef]
- Brem, S.; Grossman, S.A.; Carson, K.A.; New, P.; Phuphanich, S.; Alavi, J.B.; Mikkelsen, T.; Fisher, J.D. Phase 2 trial of copper depletion and penicillamine as antiangiogenesis therapy of glioblastoma. Neuro. Oncol. 2005, 7, 246–253. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiang, Y.; Kang, Y.J. Copper reverses cardiomyocyte hypertrophy through vascular endothelial growth factor-mediated reduction in the cell size. J. Mol. Cell. Cardiol. 2008, 45, 106–117. [Google Scholar] [CrossRef] [Green Version]
- Tardito, S.; Bassanetti, I.; Bignardi, C.; Elviri, L.; Tegoni, M.; Mucchino, C.; Bussolati, O.; Franchi-Gazzola, R.; Marchiò, L. Copper binding agents acting as copper ionophores lead to caspase inhibition and paraptotic cell death in human cancer cells. J. Am. Chem. Soc. 2011, 133, 6235–6242. [Google Scholar] [CrossRef]
- Price, K.A.; Crouch, P.J.; Volitakis, I.; Paterson, B.M.; Lim, S.; Donnelly, P.S.; White, A.R. Mechanisms controlling the cellular accumulation of copper bis(thiosemicarbazonato) complexes. Inorg. Chem. 2011, 50, 9594–9605. [Google Scholar] [CrossRef]
- Fukai, T.; Ushio-Fukai, M.; Kaplan, J.H. Copper transporters and copper chaperones: Roles in cardiovascular physiology and disease. Am. J. Physiol. Cell Physiol. 2018, 315, C186–C201. [Google Scholar] [CrossRef]
- Tardito, S.; Barilli, A.; Bassanetti, I.; Tegoni, M.; Bussolati, O.; Franchi-Gazzola, R.; Mucchino, C.; Marchiò, L. Copper-dependent cytotoxicity of 8-hydroxyquinoline derivatives correlates with their hydrophobicity and does not require caspase activation. J. Med. Chem. 2012, 55, 10448–10459. [Google Scholar] [CrossRef]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The current state of peptide drug discovery: Back to the future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Magrì, A.; Grasso, G.; Corti, F.; Finetti, F.; Greco, V.; Santoro, A.M.; Sciuto, S.; La Mendola, D.; Morbidelli, L.; Rizzarelli, E. Peptides derived from the histidine–proline rich glycoprotein bind copper ions and exhibit anti-angiogenic properties. Dalt. Trans. 2018, 47, 9492–9503. [Google Scholar] [CrossRef] [PubMed]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in signaling and disease: Beyond discovery and development. Cell. 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zilberberg, L.; Shinkaruk, S.; Lequin, O.; Rousseau, B.; Hagedorn, M.; Costa, F.; Caronzolo, D.; Balke, M.; Canron, X.; Convert, O.; et al. Structure and inhibitory effects on angiogenesis and tumor development of a new vascular endothelial growth inhibitor. J. Biol. Chem. 2003, 278, 35564–35573. [Google Scholar] [CrossRef] [Green Version]
- Vicari, D.; Foy, K.C.; Liotta, E.M.; Kaumaya, P.T.P. Engineered conformation-dependent VEGF peptide mimics are effective in inhibiting VEGF signaling pathways. J. Biol.Chem. 2011, 286, 13612–13625. [Google Scholar] [CrossRef] [Green Version]
- Mas-Moruno, C.; Rechenmacher, F.; Kessler, H. Cilengitide: The first anti-angiogenic small molecule drug candidate design, synthesis and clinical evaluation. Anticancer. Agents Med. Chem. 2010, 10, 753–768. [Google Scholar] [CrossRef] [Green Version]
- Eming, S.A.; Krieg, T. Molecular mechanisms of VEGF-A action during tissue repair. J. Investig. Dermatol. Symp. Proc. 2006, 11, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Peach, C.J.; Mignone, V.W.; Arruda, M.A.; Alcobia, D.C.; Hill, S.J.; Kilpatrick, L.E.; Woolard, J. Molecular Pharmacology of VEGF-A Isoforms: Binding and Signalling at VEGFR2. IJMS 2018, 19, 1264. [Google Scholar] [CrossRef] [Green Version]
- King, C.; Hristova, X.K. Direct measurements of VEGF–VEGFR2 binding affinities reveal the coupling between ligand binding and receptor dimerization. J. Biol. Chem. 2019, 294, 9064–9075. [Google Scholar] [CrossRef] [Green Version]
- Muller, Y.A.; Li, B.; Christinger, H.W.; Wells, J.A.; Cunningham, B.C.; de Vos, A.M. Vascular endothelial growth factor: Crystal structure and functional mapping of the kinase domain receptor binding site. Proc. Natl. Acad. Sci. USA 1997, 94, 7192–7197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacker, S.A.; Vitali, A.; Caesar, C.; Domagala, T.; Groenen, L.C.; Nice, E.; Achen, M.G.; Wilks, A.F. A mutant form of vascular endothelial growth factor (VEGF) that lacks VEGF receptor-2 activation retains the ability to induce vascular permeability. J. Biol. Chem. 1999, 274, 34884–34892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grünewald, F.S.; Prota, A.E.; Giese, A.; Ballmer-hofer, K. Structure—Function analysis of VEGF receptor activation and the role of coreceptors in angiogenic signaling. BBA—Proteins Proteom. 2010, 1804, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Grasso, G.; Santoro, A.M.; Magrì, A.; La Mendola, D.; Tomasello, M.F.; Zimbone, S.; Rizzarelli, E. The inorganic perspective of VEGF: Interactions of Cu2 + with peptides encompassing a recognition domain of the VEGF receptor. J. Inorg. Biochem. 2016, 159, 149–158. [Google Scholar] [CrossRef]
- Pilch, J.; Franzin, C.M.; Knowles, L.M.; Ferrer, F.J.; Marassi, F.M.; Ruoslahti, E. The Anti-angiogenic Peptide Anginex Disrupts the Cell Membrane. J. Mol. Biol. 2006, 356, 876–885. [Google Scholar] [CrossRef] [Green Version]
- Mayo, K.H.; J. van der Schaft, D.W.J.; Griffioen, A.W. Designed β-sheet peptides that inhibit proliferation and induce apoptosis in endothelial cells. Angiogenesis 2001, 4, 45–51. [Google Scholar] [CrossRef]
- Pannuzzo, M.; Raudino, A.; Milardi, D.; La Rosa, C.; Karttunen, M. α-Helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes. Sci. Rep. 2013, 3, 2781. [Google Scholar] [CrossRef] [Green Version]
- Sciacca, M.F.M.; Monaco, I.; La Rosa, C.; Milardi, D. The active role of Ca2+ ions in Aβ-mediated membrane damage. Chem. Commun. 2018, 54, 3629–3631. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Kotler, S.A.; Brender, J.R.; Chen, J.; Lee, D.; Ramamoorthy, A. Two-step mechanism of membrane disruption by Aβ through membrane fragmentation and pore formation. Biophys. J. 2012, 103, 702–710. [Google Scholar] [CrossRef] [Green Version]
- Marianna, F.; Tomasello, M.F.; Guarino, F.; Reina, S.; Messina, A.; De Pinto, V. The voltage-dependent anion selective channel 1 (VDAC1) topography in the mitochondrial outer membrane as detected in intact cell. PLoS ONE 2013, 8, e81522. [Google Scholar] [CrossRef]
- Kawakami, M.; Inagawa, R.; Hosokawa, T.; Saito, T.; Kurasaki, M. Mechanism of apoptosis induced by copper in PC12 cells. Food Chem. Toxicol. 2008, 46, 2157–2164. [Google Scholar] [CrossRef]
- Santo, S.; Silva, A.M.; Matos, M.; Monteiro, S.M.; Alvaro, A.M. Copper induced apoptosis in Caco-2 and Hep-G2 cells: Expression of caspases 3, 8 and 9, AIF and p53. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2016, 185–186, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, C.; Liu, Y.; Hosokawa, T.; Saito, T.; Kurasaki, M. Changes in the expression of epigenetic factors during copper-induced apoptosis in PC12 cells. J Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 2014, 49, 1023–1028. [Google Scholar] [CrossRef]
- Huang, X.; Atwood, C.S.; Moir, R.D.; Hartshorn, M.A.; Tanzi, R.E.; Bush, A.I. Trace metal contamination initiates the apparent auto-aggregation, amyloidosis and oligomerization of Alzheimer’s Ab peptides. J. Biol. Inorg. Chem. 2004, 9, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Pandini, G.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Travaglia, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The inorganic side of NGF: Copper(II) and zinc(II) affect the NGF mimicking signaling of the N-terminus peptides encompassing the recognition domain of TrkA receptor. Front. Neurosci. 2016, 10, 569. [Google Scholar] [CrossRef] [PubMed]
- Naletova, I.; Satriano, C.; Curci, A.; Margiotta, N.; Natile, G.; Arena, G.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. Cytotoxic phenanthroline derivatives alter metallostasis and redox homeostasis in neuroblastoma cells. Oncotarget 2018, 9, 36289–36316. [Google Scholar] [CrossRef] [Green Version]
- Magda, D.; Lecane, P.; Wang, Z.; Hu, W.; Thiemann, P.; Ma, X.; Dranchak, P.K.; Wang, X.; Lynch, V.; Wei, W.; et al. Synthesis and anticancer properties of water-soluble zinc ionophores. Cancer Res. 2008, 68, 5318–5325. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Zhou, Y.; Lind, S.E.; Ding, W.-Q. Clioquinol targets zinc to lysosomes in human cancer cells. Biochem. J. 2009, 417, 133–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel, K.G.; Chen, D.; Orlu, S.; Cui, Q.C.; Miller, F.R.; Dou, Q.P. Clioquinol and pyrrolidine dithiocarbamate complex with copper to form proteasome inhibitors and apoptosis inducers in human breast cancer cells. Breast Cancer Res. 2005, 7, R897–R908. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Fu, S.Y.; Wang, L.H.; Wang, F.Y.; Wang, N.N.; Cao, Q.; Wang, Y.T.; Yang, J.Y.; Wu, C.F. Copper improves the anti-angiogenic activity of disulfiram through the EGFR/Src/VEGF pathway in gliomas. Cancer Lett. 2015, 369, 86–96. [Google Scholar] [CrossRef]
- Allensworth, J.L.; Evans, M.K.; Bertucci, F.; Aldrich, A.J.; Festa, R.A.; Finetti, P.; Ueno, N.T.; Safi, R.; McDonnell, D.P.; Thiele, D.J.; et al. Disulfiram (DSF) acts as a copper ionophore to induce copper-dependent oxidative stress and mediate anti-tumor efficacy in inflammatory breast cancer. Mol. Oncol. 2015, 9, 1155–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Natale, G.; Pappalardo, G.; Milardi, D.; Sciacca, M.F.M.; Attanasio, F.; La Mendola, D.; Rizzarelli, E. Membrane interactions and conformational preferences of human and avian prion N-terminal tandem repeats: The role of copper(II) ions, pH, and membrane mimicking environments. J. Phys. Chem. B 2010, 114, 13830–13838. [Google Scholar] [CrossRef] [PubMed]
- Cansell, M.; Gouygou, J.P.; Jozefonvicz, J.; Letourneur, D. Lipid composition of cultured endothelial cells in relation to their growth. Lipids 1997, 32, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.C.M. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 1980, 104, 10–14. [Google Scholar] [CrossRef]
- Magrì, A.; Munzone, A.; Peana, M.; Medici, S.; Zoroddu, M.; Hansson, O.; Satriano, C.; Rizzarelli, E.; La Mendola, D. Coordination Environment of Cu(II) Ions Bound to N-Terminal Peptide Fragments of Angiogenin Protein. IJMS 2016, 17, 1240. [Google Scholar] [CrossRef]
- Bharathi Devi, S.R.; Dhivya, M.A.; Sulochana, K.N. Copper transporters and chaperones: Their function on angiogenesis and cellular signalling. J. Biosci. 2016, 41, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.-Q.; Lind, S.E. Metal ionophores—An emerging class of anticancer drugs. IUBMB Life 2009, 61, 1013–1018. [Google Scholar] [CrossRef] [PubMed]













| Peptide Sequences | |
|---|---|
| Ac-ESNITMQIMRIKPHQGQHIGEMSFLQHNK-NH2 | VEGF73-101 |
| Ac-ESTNIMGIMRIKPHQGQHIGEMSFLQHNK-NH2 | VEGFQ79G |
| Ac-ESTNIMQIMRGKPHQGQHIGEMSFLQHNK-NH2 | VEGFI83G |
| Ac-KPHQGQHIGEMSFLQHNK-NH2 | VEGF84-101 |
| Peptides | tR (min) |
|---|---|
| VEGF73-101 | 21.7 |
| VEGF84-101 | 18.2 |
| VEGFQ79G | 21.4 |
| VEGFI83G | 20.4 |
| pH | Ligand | UV/Vis λmax [nm] ε [M−1 cm−1]) | CD λmax [nm] (Δε[Μ−1 cm−1]) |
|---|---|---|---|
| 7 | VEGFQ79G | 581(134) | 605(−0.52) a; 514(0.75) a; 313(−0.68) b |
| 7 | VEGFI83G | 600(86) | 605(−0.34) a; 514(0.39) a; 313(−0.28) b |
| pH | Ligand | UV/Vis λmax [nm] (ε [M−1 cm−1]) | CD λmax [nm] (Δε[Μ−1 cm−1]) |
|---|---|---|---|
| 7 | VEGFQ79G | 589(207) | 605(−0.78) a; 514(0.65) a; 314(−0.44) b |
| 7 | VEGFI83G | 600(166) | 605(−0.75) a; 514(0.72) a; 314(−0.52) b |
| pH | Ligand | UV/Vis λmax [nm] (ε [M−1 cm−1]) | CD λmax [nm] (Δε[M−1 cm−1]) |
|---|---|---|---|
| 7 | VEGFQ79G | 592(244) | 606(−0.68) a; 515(0.64) a; 313(−0.41) b |
| 7 | VEGFI83G | ----- | 606(−0.83) a; 515(0.68) a; 314(−0.53) b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimbone, S.; Santoro, A.M.; La Mendola, D.; Giacomelli, C.; Trincavelli, M.L.; Tomasello, M.F.; Milardi, D.; García-Viñuales, S.; Sciacca, M.F.M.; Martini, C.; et al. The Ionophoric Activity of a Pro-Apoptotic VEGF165 Fragment on HUVEC Cells. Int. J. Mol. Sci. 2020, 21, 2866. https://doi.org/10.3390/ijms21082866
Zimbone S, Santoro AM, La Mendola D, Giacomelli C, Trincavelli ML, Tomasello MF, Milardi D, García-Viñuales S, Sciacca MFM, Martini C, et al. The Ionophoric Activity of a Pro-Apoptotic VEGF165 Fragment on HUVEC Cells. International Journal of Molecular Sciences. 2020; 21(8):2866. https://doi.org/10.3390/ijms21082866
Chicago/Turabian StyleZimbone, Stefania, Anna M. Santoro, Diego La Mendola, Chiara Giacomelli, Maria L. Trincavelli, Marianna F. Tomasello, Danilo Milardi, Sara García-Viñuales, Michele F. M. Sciacca, Claudia Martini, and et al. 2020. "The Ionophoric Activity of a Pro-Apoptotic VEGF165 Fragment on HUVEC Cells" International Journal of Molecular Sciences 21, no. 8: 2866. https://doi.org/10.3390/ijms21082866
APA StyleZimbone, S., Santoro, A. M., La Mendola, D., Giacomelli, C., Trincavelli, M. L., Tomasello, M. F., Milardi, D., García-Viñuales, S., Sciacca, M. F. M., Martini, C., & Grasso, G. (2020). The Ionophoric Activity of a Pro-Apoptotic VEGF165 Fragment on HUVEC Cells. International Journal of Molecular Sciences, 21(8), 2866. https://doi.org/10.3390/ijms21082866