The Interplay of Apoes with Syndecans in Influencing Key Cellular Events of Amyloid Pathology
Abstract
:1. Introduction
2. Results
2.1. SDCs Increase the Cellular Internalization of ApoE Isoforms
2.2. The Colocalization of SDC3 with ApoE in SH-SY5Y Cells
2.3. Overexpression of SDC3 Increases ApoE Uptake in Neurons
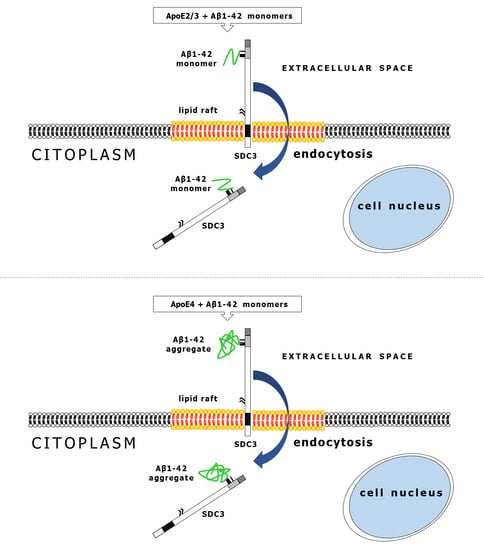
2.4. ApoEs Modulate Amyloid Pathology by Interfering with Aβ1–42 Fibrillation and Iptake
2.5. ApoEs Modulate Aβ1–42 Uptake
2.6. Colocalization of ApoE4 with Aβ1–42 Fibrils
3. Discussion
4. Materials and Methods
4.1. Peptides, Proteins and Fluorescent Labeling
4.2. SDC Constructs, Cell Culture and Transfection
4.3. Differentiation Protocol of SH-SY5Y Cells
4.4. Flow Cytometry Analysis of HS and SDC
4.5. Flow Cytometry Analysis of Cellular Uptake
4.6. Microscopic Visualization of Cellular Uptake
4.7. Co-Immunoprecipitation Experiments
4.8. Thioflavin T Binding Assays
4.9. Scanning Electron Microscopy of Surface Attachment and Fibrillation of Aβ1–42
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72 Pt A, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Mahley, R.W. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science 1988, 240, 622–630. [Google Scholar] [CrossRef]
- Tudorache, I.F.; Trusca, V.G.; Gafencu, A.V. Apolipoprotein E—A Multifunctional Protein with Implications in Various Pathologies as a Result of Its Structural Features. Comput. Struct. Biotechnol. J. 2017, 15, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Holtzman, D.M.; Herz, J.; Bu, G. Apolipoprotein E and apolipoprotein E receptors: Normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006312. [Google Scholar] [CrossRef] [Green Version]
- Elshourbagy, N.A.; Liao, W.S.; Mahley, R.W.; Taylor, J.M. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc. Natl. Acad. Sci. USA 1985, 82, 203–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahley, R.W.; Huang, Y. Apolipoprotein E: From atherosclerosis to Alzheimer’s disease and beyond. Curr. Opin. Lipidol. 1999, 10, 207–217. [Google Scholar] [CrossRef]
- Mahley, R.W.; Rall, S.C., Jr. Apolipoprotein E: Far more than a lipid transport protein. Annu. Rev. Genom. Hum. Genet. 2000, 1, 507–537. [Google Scholar] [CrossRef] [PubMed]
- Libeu, C.P.; Lund-Katz, S.; Phillips, M.C.; Wehrli, S.; Hernaiz, M.J.; Capila, I.; Linhardt, R.J.; Raffai, R.L.; Newhouse, Y.M.; Zhou, F.; et al. New insights into the heparan sulfate proteoglycan-binding activity of apolipoprotein E. J. Biol. Chem. 2001, 276, 39138–39144. [Google Scholar] [CrossRef] [Green Version]
- Leon, M.; Sawmiller, D.; Giunta, B.; Tan, J. Therapeutic approach targeting apolipoprotein E binding region and low-density lipoprotein receptor for Alzheimer’s disease. Neuroimmunol. Neuroinflammation 2018, 5, 30. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, Y.; Deguchi, N.; Takagi, C.; Tanaka, M.; Dhanasekaran, P.; Nakano, M.; Handa, T.; Phillips, M.C.; Lund-Katz, S.; Saito, H. Role of the N- and C-terminal domains in binding of apolipoprotein E isoforms to heparan sulfate and dermatan sulfate: A surface plasmon resonance study. Biochemistry 2008, 47, 6702–6710. [Google Scholar] [CrossRef] [Green Version]
- Mahley, R.W.; Ji, Z.S. Remnant lipoprotein metabolism: Key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 1999, 40, 1–16. [Google Scholar] [CrossRef]
- Altenburg, M.; Arbones-Mainar, J.; Johnson, L.; Wilder, J.; Maeda, N. Human LDL receptor enhances sequestration of ApoE4 and VLDL remnants on the surface of hepatocytes but not their internalization in mice. Arter. Thromb. Vasc. Biol. 2008, 28, 1104–1110. [Google Scholar] [CrossRef]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letoha, T.; Keller-Pinter, A.; Kusz, E.; Kolozsi, C.; Bozso, Z.; Toth, G.; Vizler, C.; Olah, Z.; Szilak, L. Cell-penetrating peptide exploited syndecans. Biochim. Biophys. Acta 2010, 1798, 2258–2265. [Google Scholar] [CrossRef] [Green Version]
- Condomitti, G.; de Wit, J. Heparan Sulfate Proteoglycans as Emerging Players in Synaptic Specificity. Front. Mol. Neurosci. 2018, 11, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letoha, T.; Hudak, A.; Kusz, E.; Pettko-Szandtner, A.; Domonkos, I.; Josvay, K.; Hofmann-Apitius, M.; Szilak, L. Contribution of syndecans to cellular internalization and fibrillation of amyloid-beta(1–42). Sci. Rep. 2019, 9, 1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]
- Park, P.W.; Reizes, O.; Bernfield, M. Cell surface heparan sulfate proteoglycans: Selective regulators of ligand-receptor encounters. J. Biol. Chem. 2000, 275, 29923–29926. [Google Scholar] [CrossRef] [Green Version]
- Hudak, A.; Kusz, E.; Domonkos, I.; Josvay, K.; Kodamullil, A.T.; Szilak, L.; Hofmann-Apitius, M.; Letoha, T. Contribution of syndecans to cellular uptake and fibrillation of alpha-synuclein and tau. Sci. Rep. 2019, 9, 16543. [Google Scholar] [CrossRef] [Green Version]
- El Masri, R.; Seffouh, A.; Lortat-Jacob, H.; Vives, R.R. The “in and out” of glucosamine 6-O-sulfation: The 6th sense of heparan sulfate. Glycoconj. J. 2017, 34, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Safaiyan, F.; Lindahl, U.; Salmivirta, M. Selective reduction of 6-O-sulfation in heparan sulfate from transformed mammary epithelial cells. Eur. J. Biochem. 1998, 252, 576–582. [Google Scholar] [CrossRef]
- Sugaya, N.; Habuchi, H.; Nagai, N.; Ashikari-Hada, S.; Kimata, K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J. Biol. Chem. 2008, 283, 10366–10376. [Google Scholar] [CrossRef] [Green Version]
- Rauch, J.N.; Chen, J.J.; Sorum, A.W.; Miller, G.M.; Sharf, T.; See, S.K.; Hsieh-Wilson, L.C.; Kampmann, M.; Kosik, K.S. Tau Internalization is Regulated by 6-O Sulfation on Heparan Sulfate Proteoglycans (HSPGs). Sci. Rep. 2018, 8, 6382. [Google Scholar] [CrossRef] [Green Version]
- Stopschinski, B.E.; Holmes, B.B.; Miller, G.M.; Manon, V.A.; Vaquer-Alicea, J.; Prueitt, W.L.; Hsieh-Wilson, L.C.; Diamond, M.I. Specific glycosaminoglycan chain length and sulfation patterns are required for cell uptake of tau versus alpha-synuclein and beta-amyloid aggregates. J. Biol. Chem. 2018, 293, 10826–10840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbull, J.; Powell, A.; Guimond, S. Heparan sulfate: Decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 2001, 11, 75–82. [Google Scholar] [CrossRef]
- Deepa, S.S.; Yamada, S.; Zako, M.; Goldberger, O.; Sugahara, K. Chondroitin sulfate chains on syndecan-1 and syndecan-4 from normal murine mammary gland epithelial cells are structurally and functionally distinct and cooperate with heparan sulfate chains to bind growth factors. A novel function to control binding of midkine, pleiotrophin, and basic fibroblast growth factor. J. Biol. Chem. 2004, 279, 37368–37376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zako, M.; Dong, J.; Goldberger, O.; Bernfield, M.; Gallagher, J.T.; Deakin, J.A. Syndecan-1 and -4 synthesized simultaneously by mouse mammary gland epithelial cells bear heparan sulfate chains that are apparently structurally indistinguishable. J. Biol. Chem. 2003, 278, 13561–13569. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, U.; Couchman, J.; Kimata, K.; Esko, J.D. Proteoglycans and Sulfated Glycosaminoglycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015; pp. 207–221. [Google Scholar]
- Kramer, K.L.; Yost, H.J. Heparan sulfate core proteins in cell-cell signaling. Annu. Rev. Genet. 2003, 37, 461–484. [Google Scholar] [CrossRef]
- Grothe, M.J.; Sepulcre, J.; Gonzalez-Escamilla, G.; Jelistratova, I.; Scholl, M.; Hansson, O.; Teipel, S.J.; Alzheimer’s Disease Neuroimaging, I. Molecular properties underlying regional vulnerability to Alzheimer’s disease pathology. Brain 2018, 141, 2755–2771. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Zhao, N.; Yamaguchi, Y.; Cirrito, J.R.; Kanekiyo, T.; Holtzman, D.M.; Bu, G. Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid-beta clearance and aggregation in Alzheimer’s disease. Sci. Transl. Med. 2016, 8, 332ra344. [Google Scholar] [CrossRef] [Green Version]
- Letoha, T.; Kolozsi, C.; Ekes, C.; Keller-pinter, A.; Kusz, E.; Szakonyi, G.; Duda, E.; Szilak, L. Contribution of syndecans to lipoplex-mediated gene delivery. Eur. J. Pharm. Sci. 2013, 49, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Hudák, A.; Letoha, A.; Szilák, L.; Letoha, T. Contribution of Syndecans to the Cellular Entry of SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 5336. [Google Scholar] [CrossRef] [PubMed]
- Afratis, N.A.; Nikitovic, D.; Multhaupt, H.A.; Theocharis, A.D.; Couchman, J.R.; Karamanos, N.K. Syndecans—Key regulators of cell signaling and biological functions. FEBS J. 2017, 284, 27–41. [Google Scholar] [CrossRef] [Green Version]
- Bernfield, M.; Kokenyesi, R.; Kato, M.; Hinkes, M.T.; Spring, J.; Gallo, R.L.; Lose, E.J. Biology of the syndecans: A family of transmembrane heparan sulfate proteoglycans. Annu. Rev. Cell Biol. 1992, 8, 365–393. [Google Scholar] [CrossRef] [PubMed]
- Lambaerts, K.; Wilcox-Adelman, S.A.; Zimmermann, P. The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr. Opin. Cell Biol. 2009, 21, 662–669. [Google Scholar] [CrossRef] [Green Version]
- Carey, D.J. Syndecans: Multifunctional cell-surface co-receptors. Biochem. J. 1997, 327 Pt 1, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Woods, A. Syndecans: Transmembrane modulators of adhesion and matrix assembly. J. Clin. Investig. 2001, 107, 935–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFall, A.J.; Rapraeger, A.C. Identification of an adhesion site within the syndecan-4 extracellular protein domain. J. Biol. Chem. 1997, 272, 12901–12904. [Google Scholar] [CrossRef] [Green Version]
- Wilsie, L.C.; Gonzales, A.M.; Orlando, R.A. Syndecan-1 mediates internalization of apoE-VLDL through a low density lipoprotein receptor-related protein (LRP)-independent, non-clathrin-mediated pathway. Lipids Health Dis. 2006, 5, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahley, R.W. Apolipoprotein E: From cardiovascular disease to neurodegenerative disorders. J. Mol. Med. 2016, 94, 739–746. [Google Scholar] [CrossRef] [Green Version]
- Namba, Y.; Tomonaga, M.; Kawasaki, H.; Otomo, E.; Ikeda, K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991, 541, 163–166. [Google Scholar] [CrossRef]
- Sundar Ganesh, H.V.; Kerman, K. Understanding the Interaction of Apolipoprotein E with Amyloid-Beta: Towards Novel Alzheimer’s Disease Therapeutics. Alzheimer’s Dement. 2016, 12, P467. [Google Scholar] [CrossRef]
- Carter, D.B. The interaction of amyloid-beta with ApoE. Subcell. Biochem. 2005, 38, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Shafti-Keramat, S.; Handisurya, A.; Kriehuber, E.; Meneguzzi, G.; Slupetzky, K.; Kirnbauer, R. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 2003, 77, 13125–13135. [Google Scholar] [CrossRef] [Green Version]
- Parolini, I.; Sargiacomo, M.; Galbiati, F.; Rizzo, G.; Grignani, F.; Engelman, J.A.; Okamoto, T.; Ikezu, T.; Scherer, P.E.; Mora, R.; et al. Expression of caveolin-1 is required for the transport of caveolin-2 to the plasma membrane. Retention of caveolin-2 at the level of the golgi complex. J. Biol. Chem. 1999, 274, 25718–25725. [Google Scholar] [CrossRef] [Green Version]
- Steinfeld, R.; van den Berghe, H.; David, G. Stimulation of fibroblast growth factor receptor-1 occupancy and signaling by cell surface-associated syndecans and glypican. J. Cell Biol. 1996, 133, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]
- Manders, E.M.; Stap, J.; Brakenhoff, G.J.; van Driel, R.; Aten, J.A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992, 103 Pt 3, 857–862. [Google Scholar] [CrossRef]
- Adler, J.; Parmryd, I. Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytom. A 2010, 77, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Kovalevich, J.; Langford, D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013, 1078, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Encinas, M.; Iglesias, M.; Liu, Y.; Wang, H.; Muhaisen, A.; Cena, V.; Gallego, C.; Comella, J.X. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 2000, 75, 991–1003. [Google Scholar] [CrossRef]
- Agholme, L.; Lindstrom, T.; Kagedal, K.; Marcusson, J.; Hallbeck, M. An in vitro model for neuroscience: Differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J. Alzheimer’s Dis. 2010, 20, 1069–1082. [Google Scholar] [CrossRef] [Green Version]
- Tsiolaki, P.L.; Katsafana, A.D.; Baltoumas, F.A.; Louros, N.N.; Iconomidou, V.A. Hidden Aggregation Hot-Spots on Human Apolipoprotein E: A Structural Study. Int. J. Mol. Sci. 2019, 20, 2274. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.C.; Zhao, N.; Fu, Y.; Wang, N.; Linares, C.; Tsai, C.W.; Bu, G. ApoE4 Accelerates Early Seeding of Amyloid Pathology. Neuron 2017, 96, 1024–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanekiyo, T.; Xu, H.; Bu, G. ApoE and Abeta in Alzheimer’s disease: Accidental encounters or partners? Neuron 2014, 81, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Frieden, C. ApoE: The role of conserved residues in defining function. Protein Sci. 2015, 24, 138–144. [Google Scholar] [CrossRef] [Green Version]
- Mahley, R.W.; Weisgraber, K.H.; Huang, Y. Apolipoprotein E: Structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 2009, 50, S183–S188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedel, B.C.; Thompson, P.M.; Brinton, R.D. Age, APOE and sex: Triad of risk of Alzheimer’s disease. J. Steroid Biochem. Mol. Biol. 2016, 160, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Safieh, M.; Korczyn, A.D.; Michaelson, D.M. ApoE4: An emerging therapeutic target for Alzheimer’s disease. BMC Med. 2019, 17, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm-Cagan, A.; Bar, R.; Harats, D.; Shaish, A.; Levkovitz, H.; Bielicki, J.K.; Johansson, J.O.; Michaelson, D.M. Differential Effects of apoE4 and Activation of ABCA1 on Brain and Plasma Lipoproteins. PLoS ONE 2016, 11, e0166195. [Google Scholar] [CrossRef] [Green Version]
- Shuvaev, V.V.; Laffont, I.; Siest, G. Kinetics of apolipoprotein E isoforms-binding to the major glycosaminoglycans of the extracellular matrix. FEBS Lett. 1999, 459, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Futamura, M.; Dhanasekaran, P.; Handa, T.; Phillips, M.C.; Lund-Katz, S.; Saito, H. Two-step mechanism of binding of apolipoprotein E to heparin: Implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J. Biol. Chem. 2005, 280, 5414–5422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerf, E.; Gustot, A.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. High ability of apolipoprotein E4 to stabilize amyloid-beta peptide oligomers, the pathological entities responsible for Alzheimer’s disease. FASEB J. 2011, 25, 1585–1595. [Google Scholar] [CrossRef]
- Blotta, S.; Jakubikova, J.; Calimeri, T.; Roccaro, A.M.; Amodio, N.; Azab, A.K.; Foresta, U.; Mitsiades, C.S.; Rossi, M.; Todoerti, K.; et al. Canonical and noncanonical Hedgehog pathway in the pathogenesis of multiple myeloma. Blood 2012, 120, 5002–5013. [Google Scholar] [CrossRef] [Green Version]
- Ellinghaus, U.; Cortini, A.; Pinder, C.L.; Le Friec, G.; Kemper, C.; Vyse, T.J. Dysregulated CD46 shedding interferes with Th1-contraction in systemic lupus erythematosus. Eur. J. Immunol. 2017, 47, 1200–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulse, K.E.; Norton, J.E.; Suh, L.; Zhong, Q.; Mahdavinia, M.; Simon, P.; Kern, R.C.; Conley, D.B.; Chandra, R.K.; Tan, B.K.; et al. Chronic rhinosinusitis with nasal polyps is characterized by B-cell inflammation and EBV-induced protein 2 expression. J. Allergy Clin. Immunol. 2013, 131, 1075–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irvin, S.C.; Herold, B.C. Molecular mechanisms linking high dose medroxyprogesterone with HIV-1 risk. PLoS ONE 2015, 10, e0121135. [Google Scholar] [CrossRef] [Green Version]
- Connor, J.P.; Felder, M.; Kapur, A.; Onujiogu, N. DcR3 binds to ovarian cancer via heparan sulfate proteoglycans and modulates tumor cells response to platinum with corresponding alteration in the expression of BRCA1. BMC Cancer 2012, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- Dieudonne, F.X.; Marion, A.; Marie, P.J.; Modrowski, D. Targeted inhibition of T-cell factor activity promotes syndecan-2 expression and sensitization to doxorubicin in osteosarcoma cells and bone tumors in mice. J. Bone Min. Res. 2012, 27, 2118–2129. [Google Scholar] [CrossRef]
- Eustace, A.D.; McNaughton, E.F.; King, S.; Kehoe, O.; Kungl, A.; Mattey, D.; Nobbs, A.H.; Williams, N.; Middleton, J. Soluble syndecan-3 binds chemokines, reduces leukocyte migration in vitro and ameliorates disease severity in models of rheumatoid arthritis. Arthritis Res. Ther. 2019, 21, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudák, A.; Jósvay, K.; Domonkos, I.; Letoha, A.; Szilák, L.; Letoha, T. The Interplay of Apoes with Syndecans in Influencing Key Cellular Events of Amyloid Pathology. Int. J. Mol. Sci. 2021, 22, 7070. https://doi.org/10.3390/ijms22137070
Hudák A, Jósvay K, Domonkos I, Letoha A, Szilák L, Letoha T. The Interplay of Apoes with Syndecans in Influencing Key Cellular Events of Amyloid Pathology. International Journal of Molecular Sciences. 2021; 22(13):7070. https://doi.org/10.3390/ijms22137070
Chicago/Turabian StyleHudák, Anett, Katalin Jósvay, Ildikó Domonkos, Annamária Letoha, László Szilák, and Tamás Letoha. 2021. "The Interplay of Apoes with Syndecans in Influencing Key Cellular Events of Amyloid Pathology" International Journal of Molecular Sciences 22, no. 13: 7070. https://doi.org/10.3390/ijms22137070
APA StyleHudák, A., Jósvay, K., Domonkos, I., Letoha, A., Szilák, L., & Letoha, T. (2021). The Interplay of Apoes with Syndecans in Influencing Key Cellular Events of Amyloid Pathology. International Journal of Molecular Sciences, 22(13), 7070. https://doi.org/10.3390/ijms22137070