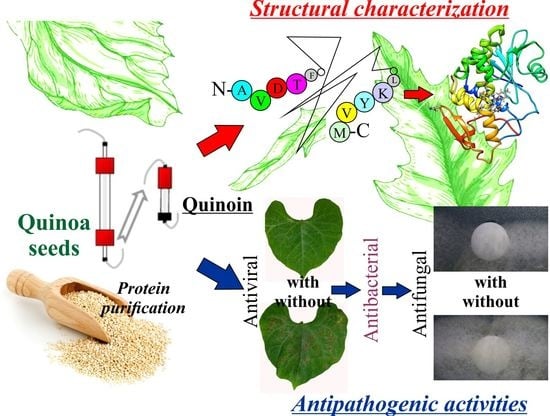
The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds
Abstract
:1. Introduction
2. Results
2.1. Protein Purification
2.2. Determination of the Primary Structure of Quinoin
2.3. Sequence Comparison, Structural Features and Homology Model of Quinoin
2.4. Gene Organization of Quinoin
2.5. Comparative MALDI-TOF Mass Spectrometry Mapping of Pre-Quinoin-1 and Pre-Quinoin-2
2.6. Antiviral Activity of Quinoin on TNV and TMV
2.7. Antimicrobial Activity
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Purification of Type 1 RIPs from the Seeds of C. quinoa
4.3. Enzymatic Assays
4.4. Analytical Procedures
4.5. Digestion and Chemical Fragmentation of Quinoin
4.6. High Resolution NanoLC-Tandem and MALDI-TOF MS Analyses
4.7. Sequence Analysis and Homology Modeling
4.8. Plant Material and Viruses for Bioassays
4.9. Evaluation of the Antiviral Activity of Quinoin In Vivo on TNV
4.10. Adenine Polynucleotide Glycosylase Activity of Quinoin on the RNA of TNV and TMV In Vitro
4.11. Evaluation of the Antibacterial Activity of Quinoin
4.12. Evaluation of the Antifungal Activity of Quinoin
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef]
- Shi, X.; Khade, P.K.; Sanbonmatsu, K.Y.; Joseph, S. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J. Mol. Biol. 2012, 419, 125–138. [Google Scholar] [CrossRef] [Green Version]
- Endo, Y.; Tsurugi, K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988, 263, 8735–8739. [Google Scholar] [CrossRef]
- De Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef] [Green Version]
- Bolognesi, A.; Polito, L.; Lubelli, C.; Barbieri, L.; Parente, A.; Stirpe, F. Ribosome-inactivating and adenine polynucleotide glycosylase activities in Mirabilis jalapa L. tissues. J. Biol. Chem. 2002, 277, 13709–13716. [Google Scholar] [CrossRef] [Green Version]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Bao, H.; Ng, T.B.; Chan, H.H.L.; Ng, C.C.W.; Man, G.C.W.; Wang, H.; Guan, S.; Zhao, S.; Fang, E.F.; et al. New ribosome-inactivating proteins and other proteins with protein synthesis-inhibiting activities. Appl. Microbiol. Biotechnol. 2020, 104, 4211–4226. [Google Scholar] [CrossRef]
- Lapadula, W.J.; Mascotti, M.L.; Juri Ayub, M. Whitefly genomes contain ribotoxin coding genes acquired from plants. Sci. Rep. 2020, 10, 15503. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tian, S.; Thaker, H.; Dong, M. Shiga Toxins: An Update on Host Factors and Biomedical Applications. Toxins 2021, 13, 222. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- Zhu, F.; Zhou, Y.-K.; Ji, Z.-L.; Chen, X.-R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-Inactivating Proteins: An Overview. In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 153–182. [Google Scholar]
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016, 23, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akkouh, O.; Ng, T.B.; Cheung, R.C.; Wong, J.H.; Pan, W.; Ng, C.C.; Sha, O.; Shaw, P.C.; Chan, W.Y. Biological activities of ribosome-inactivating proteins and their possible applications as antimicrobial, anticancer, and anti-pest agents and in neuroscience research. Appl. Microbiol. Biotechnol. 2015, 99, 9847–9863. [Google Scholar] [CrossRef]
- Citores, L.; Iglesias, R.; Ferreras, J.M. Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80. [Google Scholar] [CrossRef]
- Bulgari, D.; Landi, N.; Ragucci, S.; Faoro, F.; Di Maro, A. Antiviral Activity of PD-L1 and PD-L4, Type 1 Ribosome Inactivating Proteins from Leaves of Phytolacca dioica L. in the Pathosystem Phaseolus vulgaris-Tobacco Necrosis Virus (TNV). Toxins 2020, 12, 524. [Google Scholar] [CrossRef]
- Barbieri, L.; Polito, L.; Bolognesi, A.; Ciani, M.; Pelosi, E.; Farini, V.; Jha, A.K.; Sharma, N.; Vivanco, J.M.; Chambery, A.; et al. Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta 2006, 1760, 783–792. [Google Scholar] [CrossRef] [PubMed]
- De Zaeytijd, J.; Van Damme, E.J. Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles. Toxins 2017, 9, 123. [Google Scholar] [CrossRef]
- Chen, Y.J.; Zhu, J.Q.; Fu, X.Q.; Su, T.; Li, T.; Guo, H.; Zhu, P.L.; Lee, S.K.; Yu, H.; Tse, A.K.; et al. Ribosome-Inactivating Protein α-Momorcharin Derived from Edible Plant Momordica charantia Induces Inflammatory Responses by Activating the NF-kappaB and JNK Pathways. Toxins 2019, 11, 694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barre, A.; Damme, E.; Simplicien, M.; Benoist, H.; Rougé, P. Are Dietary Lectins Relevant Allergens in Plant Food Allergy? Foods 2020, 9, 1724. [Google Scholar] [CrossRef]
- Landi, N.; Ruocco, M.R.; Ragucci, S.; Aliotta, F.; Nasso, R.; Pedone, P.V.; Di Maro, A. Quinoa as source of type 1 ribosome inactivating proteins: A novel knowledge for a revision of its consumption. Food Chem. 2021, 342, 128337. [Google Scholar] [CrossRef]
- Filho, A.M.; Pirozi, M.R.; Borges, J.T.; Pinheiro Sant’Ana, H.M.; Chaves, J.B.; Coimbra, J.S. Quinoa: Nutritional, functional, and antinutritional aspects. Crit. Rev. Food Sci. Nutr. 2017, 57, 1618–1630. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Cooper, C.A.; Gasteiger, E.; Packer, N.H. GlycoMod -a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 2001, 1, 340–349. [Google Scholar] [CrossRef]
- Takahashi, N.; Lee, K.B.; Nakagawa, H.; Tsukamoto, Y.; Masuda, K.; Lee, Y.C. New N-glycans in horseradish peroxidase. Anal. Biochem. 1998, 255, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.B.; Zeleny, R.; Kolarich, D.; Staudacher, E.; Stroop, C.J.; Kamerling, J.P.; Altmann, F. Analysis of Asn-linked glycans from vegetable foodstuffs: Widespread occurrence of Lewis a, core alpha1,3-linked fucose and xylose substitutions. Glycobiology 2001, 11, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K. Enzymes: An integrated view of structure, dynamics and function. Microb. Cell Fact. 2006, 5, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambery, A.; Pisante, M.; Di Maro, A.; Di Zazzo, E.; Ruvo, M.; Costantini, S.; Colonna, G.; Parente, A. Invariant Ser211 is involved in the catalysis of PD-L4, type I RIP from Phytolacca dioica leaves. Proteins 2007, 67, 209–218. [Google Scholar] [CrossRef]
- Li, X.P.; Kahn, P.C.; Kahn, J.N.; Grela, P.; Tumer, N.E. Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J. Biol. Chem. 2013, 288, 30270–30284. [Google Scholar] [CrossRef] [Green Version]
- Chambery, A.; Di Maro, A.; Parente, A. Primary structure and glycan moiety characterization of PD-Ss, type 1 ribosome-inactivating proteins from Phytolacca dioica L. seeds, by precursor ion discovery on a Q-TOF mass spectrometer. Phytochemistry 2008, 69, 1973–1982. [Google Scholar] [CrossRef]
- Di Maro, A.; Valbonesi, P.; Bolognesi, A.; Stirpe, F.; De Luca, P.; Siniscalco Gigliano, G.; Gaudio, L.; Delli Bovi, P.; Ferranti, P.; Malorni, A.; et al. Isolation and characterization of four type-1 ribosome-inactivating proteins, with polynucleotide:adenosine glycosidase activity, from leaves of Phytolacca dioica L. Planta 1999, 208, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Robertus, J.D.; Monzingo, A.F. The structure of ribosome inactivating proteins. Mini Rev. Med. Chem. 2004, 4, 477–486. [Google Scholar] [CrossRef]
- Reyes, L.F.; Nobre, T.M.; Pavinatto, F.J.; Zaniquelli, M.E.; Caseli, L.; Oliveira, O.N., Jr.; Araújo, A.P. The role of the C-terminal region of pulchellin A-chain in the interaction with membrane model systems. Biochim. Biophys. Acta 2012, 1818, 82–89. [Google Scholar] [CrossRef]
- He, W.J.; Liu, W.Y. Both N- and C-terminal regions are essential for cinnamomin A-chain to deadenylate ribosomal RNA and supercoiled double-stranded DNA. Biochem. J. 2004, 377, 17–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baykal, U.; Tumer, N.E. The C-terminus of pokeweed antiviral protein has distinct roles in transport to the cytosol, ribosome depurination and cytotoxicity. Plant J. 2007, 49, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Ragucci, S.; Russo, R.; Di Maro, A.; Ferreras, J.M. Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L. Biochim. Biophys. Acta 2016, 1860, 1256–1264. [Google Scholar] [CrossRef]
- Stirpe, F.; Barbieri, L.; Gorini, P.; Valbonesi, P.; Bolognesi, A.; Polito, L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996, 382, 309–312. [Google Scholar] [CrossRef] [Green Version]
- Wytynck, P.; Lambin, J.; Chen, S.; Demirel Asci, S.; Verbeke, I.; De Zaeytijd, J.; Subramanyam, K.; Van Damme, E.J.M. Effect of RIP Overexpression on Abiotic Stress Tolerance and Development of Rice. Int. J. Mol. Sci. 2021, 22, 1434. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Mercatelli, D.; Mancuso, R.; Baruzzi, G.; Faedi, W.; Bolognesi, A. Protein synthesis inhibition activity by strawberry tissue protein extracts during plant life cycle and under biotic and abiotic stresses. Int. J. Mol. Sci. 2013, 14, 15532–15545. [Google Scholar] [CrossRef] [PubMed]
- Neller, K.C.M.; Diaz, C.A.; Platts, A.E.; Hudak, K.A. De novo Assembly of the Pokeweed Genome Provides Insight into Pokeweed Antiviral Protein (PAP) Gene Expression. Front. Plant Sci. 2019, 10, 1002. [Google Scholar] [CrossRef] [Green Version]
- Barbieri, L.; Valbonesi, P.; Gorini, P.; Pession, A.; Stirpe, F. Polynucleotide: Adenosine glycosidase activity of saporin-L1: Effect on DNA, RNA and poly(A). Biochem. J. 1996, 319, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Vivanco, J.M.; Savary, B.J.; Flores, H.E. Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the andean crop Mirabilis expansa. Plant Physiol. 1999, 119, 1447–1456. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.; Park, S.W.; Vepachedu, R.; Barbieri, L.; Ciani, M.; Stirpe, F.; Savary, B.J.; Vivanco, J.M. Isolation and characterization of an RIP (ribosome-inactivating protein)-like protein from tobacco with dual enzymatic activity. Plant Physiol. 2004, 134, 171–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezaei-Moshaei, M.; Dehestani, A.; Bandehagh, A.; Pakdin-Parizi, A.; Golkar, M.; Heidari-Japelaghi, R. Recombinant pebulin protein, a type 2 ribosome-inactivating protein isolated from dwarf elder (Sambucus ebulus L.) shows anticancer and antifungal activities in vitro. Int. J. Biol. Macromol. 2021, 174, 352–361. [Google Scholar] [CrossRef]
- Aurich, S.; Simon, J.C.; Treudler, R. New and rare elicitors of food allergy. Allergologie 2019, 42, 83–90. [Google Scholar] [CrossRef]
- Zevallos, V.F.; Ellis, H.J.; Suligoj, T.; Herencia, L.I.; Ciclitira, P.J. Variable activation of immune response by quinoa (Chenopodium quinoa Willd.) prolamins in celiac disease. Am. J. Clin. Nutr. 2012, 96, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Maro, A.; Terracciano, I.; Sticco, L.; Fiandra, L.; Ruocco, M.; Corrado, G.; Parente, A.; Rao, R. Purification and characterization of a viral chitinase active against plant pathogens and herbivores from transgenic tobacco. J. Biotechnol. 2010, 147, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Landi, N.; Pacifico, S.; Ragucci, S.; Iglesias, R.; Piccolella, S.; Amici, A.; Di Giuseppe, A.M.A.; Di Maro, A. Purification, characterization and cytotoxicity assessment of Ageritin: The first ribotoxin from the basidiomycete mushroom Agrocybe aegerita. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt A, 1113–1121. [Google Scholar] [CrossRef]
- Di Maro, A.; Ferranti, P.; Mastronicola, M.; Polito, L.; Bolognesi, A.; Stirpe, F.; Malorni, A.; Parente, A. Reliable sequence determination of ribosome-inactivating proteins by combining electrospray mass spectrometry and Edman degradation. J. Mass. Spectrom. 2001, 36, 38–46. [Google Scholar] [CrossRef]
- Landi, N.; Ragucci, S.; Russo, R.; Pedone, P.V.; Chambery, A.; Di Maro, A. Structural insights into nucleotide and protein sequence of Ageritin: A novel prototype of fungal ribotoxin. J. Biochem. 2019, 165, 415–422. [Google Scholar] [CrossRef]
- Di Giuseppe, A.M.A.; Russo, L.; Russo, R.; Ragucci, S.; Caso, J.V.; Isernia, C.; Chambery, A.; Di Maro, A. Molecular characterization of myoglobin from Sciurus vulgaris meridionalis: Primary structure, kinetics and spectroscopic studies. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gooding, G.V., Jr.; Hebert, T.T. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 1967, 57, 1285. [Google Scholar]
- Montalbini, P.; Polverari, A. Characterization of a tobacco necrosis virus strain isolated from tomato leaves. Phytopathol. Mediterr. 1996, 35, 105–110. [Google Scholar]
- Baer, A.; Kehn-Hall, K. Viral concentration determination through plaque assays: Using traditional and novel overlay systems. J. Vis. Exp. 2014, 93, e52065. [Google Scholar] [CrossRef]
- Schlumbaum, A.; Mauch, F.; Vögeli, U.; Boller, T. Plant chitinases are potent inhibitors of fungal growth. Nature 1986, 324, 365–367. [Google Scholar] [CrossRef]





| Sequence | MC | Charge | m/z [Da] | MH+ [Da] | Δmass [ppm] | Sequence Position | Reported in Figure 2 |
|---|---|---|---|---|---|---|---|
| A | |||||||
| [R].NDLYVVAFADK.[F] | - | 2 | 627.822 | 1254.636 | 0.45 | 72–82 | TMS/MS-1 |
| [R].GHFFSNLNIDTIDK.[A] | - | 3 | 540.938 | 1620.801 | 0.59 | 89–102 | TMS/MS-2 |
| [R].LSFPLGFDNLK.[T] | - | 2 | 625.842 | 1250.677 | 0.72 | 134–144 | TMS/MS-3 |
| [K].VYGMDTK.[A] | - | 2 | 407.194 | 829.381 | 0.56 | 150–156 | TMS/MS-4 |
| [R].AIVTTNPNNYK.[I] | - | 2 | 617.824 | 1234.64 | 0.59 | 188–198 | TMS/MS-5 |
| [K].ILSLENNWGAISK.[G] | - | 2 | 772.892 | 1444.78 | 1.01 | 199–211 | TMS/MS-6 |
| [K].NDMGLLK.[Y] | - | 2 | 395.710 | 790.412 | 0.44 | 245–251 | TMS/MS-7 |
| B | |||||||
| [K].LEPKPTQNTY.[N] | - | 2 | 595.806 | 1190.604 | 0.64 | 7–16 | n.r. |
| [K].LEPKPTQNTYNTF.[L] | Y16 | 2 | 776.885 | 1552.763 | 1.02 | 7–19 | ChMS/MS-1 |
| [K].DPSLVYEGIPM.[I] | - | 2 | 610.797 | 1220.586 | 0.73 | 29–39 | ChMS/MS-2 |
| [Y].LLVDLESK.[K] | - | 2 | 458.770 | 916.534 | 0.81 | 50–57 | ChMS/MS-3 |
| Peptide | Sequence Position | Experimental Molecular Mass | Theoretical Molecular Mass a | Δ(Da) | Missed Cleavage at | Reported in Figure 2 |
|---|---|---|---|---|---|---|
| Arg-C peptides | ||||||
| 1 | 1–24 | 2811.68 | 2811.47 | 0.21 | - | Arg-1 |
| 2 | 25–41 | 1934.75 | 1934.00 | 0.75 | - | Arg-2 |
| chymotrypsin peptides | ||||||
| 1 | 40–49 | 1173.57 | 1173.59 | 0.02 | - | Ch-1 |
| 2 | 50–64 | 1791.26 | 1790.99 | 0.27 | - | Ch-2 |
| 3 | 84–91 | 857.48 | 857.47 | 0.01 | - | Ch-3 |
| 4 | 93–103 | 2405.82 | 2405.31 | 0.51 | - | Ch-4 |
| trypsin peptides | ||||||
| 1 | 59–71 | 1505.68 | 1505.83 | 0.15 | K60 | T-1 |
| 2 | 72–86 | 1643.64 | 1643.84 | 0.20 | K82 | T-2 |
| 3 | 103–133 | 3489.78 | 3489.76 | 0.02 | K104, K105 | T-3 |
| 4 | 167–187 | 2438.91 | 2438.37 | 0.54 | R181, K183 | T-4 |
| 5 | 199–219 | 2311.03 | 2311.32 | 0.29 | K211, R214, K218 | T-5 |
| 6 | 220–244 | 3490.28 | 3487.87 b | 2.41 | - | T-6 |
| 6′ | 220–244 | 3636.56 | 3635.99 c | 0.57 | - | (T-6) |
| CNBr fragments | ||||||
| 1 | 148–153 | 678.36 | 678.35 d | 0.01 | - | CB-1 |
| 2 | 154–174 | 2394.34 | 2394.30 d | 0.04 | - | CB-2 |
| 3 | 248–254 | 793.46 | 793.00 d | 0.46 | - | CB-3 |
| Theoretical Molecular Masses | Sequence Position | Experimental Molecular Masses a | Missed Cleavage | |||
|---|---|---|---|---|---|---|
| Pre-Quinoin-1 | Δ(Da) | Pre-Quinoin-2 | Δ(Da) | |||
| 790.41 | 245–251 | 790.72 | 0.31 | 790.73 | 0.32 | - |
| 813.64 | 150–156 | 813.66 | 0.02 | 813.68 | 0.04 | - |
| 1234.64 | 188–198 | 1234.53 | 0.11 | 1234.58 | 0.06 | - |
| 1250.67 | 134–144 | 1250.56 | 0.11 | 1250.60 | 0.07 | - |
| 1254.63 | 72–82 | 1254.51 | 0.12 | 1254.58 | 0.05 | - |
| 1444.77 | 199–211 | 1444.45 | 0.32 | 1444.53 | 0.24 | - |
| 1620.80 | 98–102 | 1620.32 | 0.48 | 1620.28 | 0.52 | - |
| 1643.84 | 72–86 | 1643.33 | 0.51 | n.r. | K82 | |
| 1664.80 | 145–158 | 1664.30 | 0.50 | n.r. | K149, K156 | |
| 1819.93 | 89–104 | 1819.31 | 0.62 | n.r. | K102 | |
| 2150.12 | 7–24 | 2149.27 | 0.85 | 2149.31 | 0.81 | - |
| 2278.27 | 163–182 | 2277.31 | 0.96 | n.r. | R166, R180 | |
| 2311.32 | 199–219 | 2311.29 | 0.03 | n.r. | K211, R214, K218 | |
| 2438.37 | 167–187 | 2439.34 | 0.97 | 2439.11 | 0.74 | R180, K182 |
| Microbe | Zone of Inhibition (mm ± SD) | ||
|---|---|---|---|
| Protein Amount | |||
| 0.2 µg | 0.5 µg | 1.0 µg | |
| Bacteria | |||
| Pseudomonas syringae pv phaseolicola | 2.5 ± 0.3 | 3.4 ± 0.5 | 4.0 ± 0.5 |
| Pseudomonas syringae pv actinidiae | 1.5 ± 0.3 | 1.8 ± 0.3 | 2.0 ± 0.4 |
| Salmonella enterica | 0.0 | 0.0 | 0.0 |
| Staphylococcus aureus | 0.0 | 0.0 | 0.0 |
| Fungi | |||
| Cryponectria parasitica strain E4 | 0.0 | 0.0 | 0.0 |
| Cryponectria parasitica strain E13 | 0.0 | 0.0 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragucci, S.; Bulgari, D.; Landi, N.; Russo, R.; Clemente, A.; Valletta, M.; Chambery, A.; Gobbi, E.; Faoro, F.; Di Maro, A. The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds. Int. J. Mol. Sci. 2021, 22, 8964. https://doi.org/10.3390/ijms22168964
Ragucci S, Bulgari D, Landi N, Russo R, Clemente A, Valletta M, Chambery A, Gobbi E, Faoro F, Di Maro A. The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds. International Journal of Molecular Sciences. 2021; 22(16):8964. https://doi.org/10.3390/ijms22168964
Chicago/Turabian StyleRagucci, Sara, Daniela Bulgari, Nicola Landi, Rosita Russo, Angela Clemente, Mariangela Valletta, Angela Chambery, Emanuela Gobbi, Franco Faoro, and Antimo Di Maro. 2021. "The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds" International Journal of Molecular Sciences 22, no. 16: 8964. https://doi.org/10.3390/ijms22168964
APA StyleRagucci, S., Bulgari, D., Landi, N., Russo, R., Clemente, A., Valletta, M., Chambery, A., Gobbi, E., Faoro, F., & Di Maro, A. (2021). The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds. International Journal of Molecular Sciences, 22(16), 8964. https://doi.org/10.3390/ijms22168964