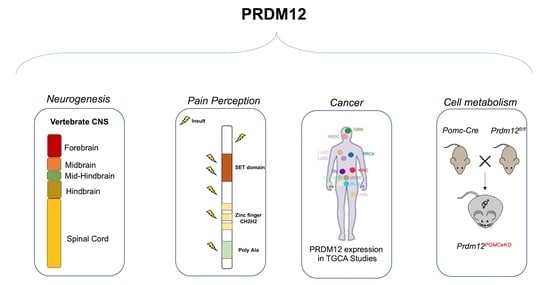
PRDM12 in Health and Diseases
Abstract
:1. Introduction
2. PRDM12 Gene and Its Protein Product
3. Established PRDM12 Functions: Neurogenesis
4. Established PRDM12 Functions: Pain Perception
5. Exploring Novel PRDM12 Functions: Cancer
6. Exploring Novel PRDM12 Functions: Cell Metabolism
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Casamassimi, A.; Rienzo, M.; Di Zazzo, E.; Sorrentino, A.; Fiore, D.; Proto, M.C.; Moncharmont, B.; Gazzerro, P.; Bifulco, M.; Abbondanza, C. Multifaceted Role of PRDM Proteins in Human Cancer. Int. J. Mol. Sci. 2020, 21, 2648. [Google Scholar] [CrossRef] [Green Version]
- Di Zazzo, E.; De Rosa, C.; Abbondanza, C.; Moncharmont, B. PRDM Proteins: Molecular Mechanisms in Signal Transduction and Transcriptional Regulation. Biology 2013, 2, 107–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef]
- Eram, M.S.; Bustos, S.P.; Fernandes, E.L.; Siarheyeva, A.; Senisterra, G.; Hajian, T.; Chau, I.; Duan, S.; Wu, H.; Dombrovski, L.; et al. Trimethylation of Histone H3 Lysine 36 by Human Methyltransferase PRDM9 Protein. J. Biol. Chem. 2014, 289, 12177–12188. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 Methyltransferases Required for Mammalian Heterochromatin Integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zannino, D.A.; Sagerström, C.G. An emerging role for prdm family genes in dorsoventral patterning of the vertebrate nervous system. Neural Dev. 2015, 10, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Shao, G.; Liu, L. The PR Domain of the Rb-binding Zinc Finger Protein RIZ1 Is a Protein Binding Interface and Is Related to the SET Domain Functioning in Chromatin-mediated Gene Expression. J. Biol. Chem. 1998, 273, 15933–15939. [Google Scholar] [CrossRef] [Green Version]
- Ren, B.; Chee, K.J.; Kim, T.H.; Maniatis, T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999, 13, 125–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okashita, N.; Suwa, Y.; Nishimura, O.; Sakashita, N.; Kadota, M.; Nagamatsu, G.; Kawaguchi, M.; Kashida, H.; Nakajima, A.; Tachibana, M.; et al. PRDM14 Drives OCT3/4 Recruitment via Active Demethylation in the Transition from Primed to Naive Pluripotency. Stem Cell Rep. 2016, 7, 1072–1086. [Google Scholar] [CrossRef] [Green Version]
- Chu, L.-F.; Surani, A.; Jaenisch, R.; Zwaka, T.P. Blimp1 Expression Predicts Embryonic Stem Cell Development in Vitro. Curr. Biol. 2011, 21, 1759–1765. [Google Scholar] [CrossRef] [Green Version]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, J.; Cohen, P. The Multifaceted Roles of PRDM16: Adipose Biology and Beyond. Trends Endocrinol. Metab. 2016, 27, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Lagunas, L.; Choi, I.F.; Kaji, T.; Simpson, P.; Hershey, C.; Zhou, Y.; Zon, L.; Mercola, M.; Artinger, K.B. Zebrafish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev. Biol. 2005, 278, 347–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komai, T.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Shinkai, Y. Expression of the mouse PR domain protein Prdm8 in the developing central nervous system. Gene Expr. Patterns 2009, 9, 503–514. [Google Scholar] [CrossRef]
- Ross, S.E.; McCord, A.E.; Jung, C.; Atan, D.; Mok, S.I.; Hemberg, M.; Kim, T.-K.; Salogiannis, J.; Hu, L.; Cohen, S.; et al. Bhlhb5 and Prdm8 Form a Repressor Complex Involved in Neuronal Circuit Assembly. Neuron 2012, 73, 292–303. [Google Scholar] [CrossRef] [Green Version]
- Inoue, M.; Iwai, R.; Tabata, H.; Konno, D.; Komabayashi-Suzuki, M.; Watanabe, C.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Matsuzaki, F.; et al. Prdm16 is critical for progression of the multipolar phase during neural differentiation of the developing neocortex. Development 2017, 144, 385–399. [Google Scholar] [CrossRef] [Green Version]
- Shimada, I.S.; Acar, M.; Burgess, R.J.; Zhao, Z.; Morrison, S.J. Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells. Genes Dev. 2017, 31, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Baizabal, J.-M.; Mistry, M.; García, M.T.; Gómez, N.; Olukoya, O.; Tran, D.; Johnson, M.B.; Walsh, C.A.; Harwell, C.C. The Epigenetic State of PRDM16-Regulated Enhancers in Radial Glia Controls Cortical Neuron Position. Neuron 2018, 98, 945–962.e8. [Google Scholar] [CrossRef] [Green Version]
- Kinameri, E.; Inoue, T.; Aruga, J.; Imayoshi, I.; Kageyama, R.; Shimogori, T.; Moore, A.W. Prdm Proto-Oncogene Transcription Factor Family Expression and Interaction with the Notch-Hes Pathway in Mouse Neurogenesis. PLoS ONE 2008, 3, e3859. [Google Scholar] [CrossRef] [Green Version]
- Woo, J.; Kang, H.; Lee, E.Y.; Park, S.; Cho, Y.E. Investigation of PRDM7 and PRDM12 expression pattern during mouse embryonic development by using a modified passive clearing technique. Biochem. Biophys. Res. Commun. 2020, 524, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Kim, I.; Ahn, D.; Tae, H.; Park, B. PR domaincontaining protein 12 (prdm12) is a downstream target of the transcription factor zic1 during cellular differentiation in the central nervous system: PR domain containing protein is the right form. Int. J. Dev. Neurosci. 2020, 80, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Hael, C.E.; Rojo, D.; Orquera, D.P.; Low, M.J.; Rubinstein, M. The transcriptional regulator PRDM12 is critical for Pomc expression in the mouse hypothalamus and controlling food intake, adiposity, and body weight. Mol. Metab. 2020, 34, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Chittka, A.; Nitarska, J.; Grazini, U.; Richardson, W.D. Transcription Factor Positive Regulatory Domain 4 (PRDM4) Recruits Protein Arginine Methyltransferase 5 (PRMT5) to Mediate Histone Arginine Methylation and Control Neural Stem Cell Proliferation and Differentiation. J. Biol. Chem. 2012, 287, 42995–43006. [Google Scholar] [CrossRef] [Green Version]
- Hadziselimovic, F.; Cathomas, G.; Verkauskas, G.; Dasevicius, D.; Stadler, M.B. PRDM Histone Methyltransferase mRNA Levels Increase in Response to Curative Hormone Treatment for Cryptorchidism-Dependent Male Infertility. Genes 2018, 9, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrentino, A.; Federico, A.; Rienzo, M.; Gazzerro, P.; Bifulco, M.; Ciccodicola, A.; Casamassimi, A.; Abbondanza, C. PR/SET Domain Family and Cancer: Novel Insights from the Cancer Genome Atlas. Int. J. Mol. Sci. 2018, 19, 3250. [Google Scholar] [CrossRef] [Green Version]
- Di Tullio, F.; Schwarz, M.; Zorgati, H.; Mzoughi, S.; Guccione, E. The duality of PRDM proteins: Epigenetic and structural perspectives. FEBS J. 2021. [Google Scholar] [CrossRef]
- Rienzo, M.; Sorrentino, A.; Di Zazzo, E.; Di Donato, M.; Carafa, V.; Marino, M.M.; De Rosa, C.; Gazzerro, P.; Castoria, G.; Altucci, L.; et al. Searching for a Putative Mechanism of RIZ2 Tumor-Promoting Function in Cancer Models. Front. Oncol. 2021, 10, 583533. [Google Scholar] [CrossRef]
- Mzoughi, S.; Tan, Y.X.; Low, D.; Guccione, E. The role of PRDMs in cancer: One family, two sides. Curr. Opin. Genet. Dev. 2016, 36, 83–91. [Google Scholar] [CrossRef]
- Dettman, E.; Simko, S.J.; Ayanga, B.; Carofino, B.; Margolin, J.; Morse, H.; Justice, M.J. Prdm14 initiates lymphoblastic leukemia after expanding a population of cells resembling common lymphoid progenitors. Oncogene 2011, 30, 2859–2873. [Google Scholar] [CrossRef] [Green Version]
- Mzoughi, S.; Fong, J.Y.; Papadopoli, D.; Koh, C.M.; Hulea, L.; Pigini, P.; Di Tullio, F.; Andreacchio, G.; Hoppe, M.M.; Wollmann, H.; et al. PRDM15 is a key regulator of metabolism critical to sustain B-cell lymphomagenesis. Nat. Commun. 2020, 11, 3520. [Google Scholar] [CrossRef]
- Imhof, S.; Kokotović, T.; Nagy, V. PRDM12: New Opportunity in Pain Research. Trends Mol. Med. 2020, 26, 895–897. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Auer-Grumbach, M.; Matsukawa, S.; Zitzelsberger, M.; Themistocleous, A.; Strom, T.M.; Samara, C.; Moore, A.W.; Cho, L.T.-Y.; Young, G.T.; et al. Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 2015, 47, 803–808. [Google Scholar] [CrossRef]
- Available online: https://www.proteinatlas.org/ENSG00000130711-PRDM12/cell (accessed on 1 April 2021).
- Yang, C.-M.; Shinkai, Y. Prdm12 Is Induced by Retinoic Acid and Exhibits Anti-proliferative Properties through the Cell Cycle Modulation of P19 Embryonic Carcinoma Cells. Cell Struct. Funct. 2013, 38, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Thélie, A.; Desiderio, S.; Hanotel, J.; Quigley, I.; Van Driessche, B.; Rodari, A.; Borromeo, M.D.; Kricha, S.; Lahaye, F.; Croce, J.; et al. Prdm12 specifies V1 interneurons through cross-repressive interactions with Dbx1 and Nkx6 genes in Xenopus. Development 2015, 142, 3416–3428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yildiz, O.; Downes, G.B.; Sagerström, C.G. Zebrafish prdm12b acts independently of nkx6.1 repression to promote eng1b expression in the neural tube p1 domain. Neural Dev. 2019, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, S.; Miwata, K.; Asashima, M.; Michiue, T. The requirement of histone modification by PRDM12 and Kdm4a for the development of pre-placodal ectoderm and neural crest in Xenopus. Dev. Biol. 2015, 399, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Vervoort, M.; Meulemeester, D.; Béhague, J.; Kerner, P. Evolution of Prdm Genes in Animals: Insights from Comparative Genomics. Mol. Biol. Evol. 2015, 33, 679–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.ncbi.nlm.nih.gov/gene/59335/ortholog/?scope=89593&term=PRDM12 (accessed on 1 April 2021).
- Zannino, D.A.; Downes, G.B.; Sagerström, C.G. prdm12b specifies the p1 progenitor domain and reveals a role for V1 interneurons in swim movements. Dev. Biol. 2014, 390, 247–260. [Google Scholar] [CrossRef] [Green Version]
- Gard, C.; Curto, G.G.; Frarma, Y.E.-M.; Chollet, E.; Duval, N.; Auzié, V.; Auradé, F.; Vigier, L.; Relaix, F.; Pierani, A.; et al. Pax3- and Pax7-mediated Dbx1 regulation orchestrates the patterning of intermediate spinal interneurons. Dev. Biol. 2017, 432, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Mona, B.; Uruena, A.; Kollipara, R.K.; Ackerman, S.L.; Borromeo, M.D.; Chang, J.C.; Johnson, J.E. Repression by PRDM13 is critical for generating precision in neuronal identity. eLife 2017, 6, e25787. [Google Scholar] [CrossRef]
- Wang, J.; Kollarik, M.; Ru, F.; Sun, H.; McNeil, B.; Dong, X.; Stephens, G.; Korolevich, S.; Brohawn, P.; Kolbeck, R.; et al. Distinct and common expression of receptors for inflammatory mediators in vagal nodose versus jugular capsaicin-sensitive/TRPV1-positive neurons detected by low input RNA sequencing. PLoS ONE 2017, 12, e0185985. [Google Scholar] [CrossRef] [Green Version]
- Iacono, L.L.; Valzania, A.; Visco-Comandini, F.; Viscomi, M.T.; Felsani, A.; Puglisi-Allegra, S.; Carola, V. Regulation of nucleus accumbens transcript levels in mice by early-life social stress and cocaine. Neuropharmacology 2016, 103, 183–194. [Google Scholar] [CrossRef]
- Nahorski, M.S.; Chen, Y.-C.; Woods, C.G. New Mendelian Disorders of Painlessness. Trends Neurosci. 2015, 38, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Drissi, I.; Woods, W.A.; Woods, C.G. Understanding the genetic basis of congenital insensitivity to pain. Br. Med. Bull. 2020, 133, 65–78. [Google Scholar] [CrossRef]
- Nagy, V.V.; Cole, T.T.; Van Campenhout, C.; Khoung, T.T.; Leung, C.C.; Vermeiren, S.S.; Novatchkova, M.M.; Wenzel, D.D.; Cikes, D.D.; Polyansky, A.A.; et al. The evolutionarily conserved transcription factor PRDM12 controls sensory neuron development and pain perception. Cell Cycle 2015, 14, 1799–1808. [Google Scholar] [CrossRef]
- Zhang, S.; Sharif, S.M.; Chen, Y.-C.; Valente, E.-M.; Ahmed, M.; Sheridan, E.; Bennett, C.; Woods, G. Clinical features for diagnosis and management of patients with PRDM12 congenital insensitivity to pain. J. Med. Genet. 2016, 53, 533–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, A.G.; Padmanabh, H.; Sahu, J.K.; Kurth, I.; Voigt, M.; Singhi, P. Hereditary Sensory Polyneuropathy, Pain Insensitivity and Global Developmental Delay due to Novel Mutation in PRDM12 Gene. Indian J. Pediatr. 2017, 84, 332–333. [Google Scholar] [CrossRef]
- Elhennawy, K.; Reda, S.; Finke, C.; Graul-Neumann, L.; Jost-Brinkmann, P.-G.; Bartzela, T. Oral manifestations, dental management, and a rare homozygous mutation of the PRDM12 gene in a boy with hereditary sensory and autonomic neuropathy type VIII: A case report and review of the literature. J. Med. Case Rep. 2017, 11, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivas, S.M.; Gowda, V.K.; Owen, C.M.; Moss, C.; Hiremagalore, R. Mid-face toddler excoriation syndrome (MiTES): A new paediatric diagnosis. Clin. Exp. Dermatol. 2017, 42, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Moss, C.; Srinivas, S.M.; Sarveswaran, N.; Nahorski, M.; Gowda, V.K.; Browne, F.M.; Woods, G. Midface toddler excoriation syndrome (MiTES) can be caused by autosomal recessive biallelic mutations in a gene for congenital insensitivity to pain, PRDM12. Br. J. Dermatol. 2018, 179, 1135–1140. [Google Scholar] [CrossRef]
- Inamadar, A.C.; Vinay, K.; Olabi, B.; Sarveswaran, N.; Bishnoi, A.; Woods, C.G.; Moss, C. Extending the phenotype of midface toddler excoriation syndrome (MiTES): Five new cases in three families with PR domain containing protein 12 (PRDM12) mutations. J. Am. Acad. Dermatol. 2019, 81, 1415–1417. [Google Scholar] [CrossRef]
- Navya, M.K.; Pramod, G.V.; Sujatha, G.P.; Ashok, L. Congenital insensitivity to pain in a 1-year-old boy. J. Indian Soc. Pedod. Prev. Dent. 2019, 37, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Noguera-Morel, L.; Ortiz-Cabrera, N.V.; Campos, M.; Hernández-Martín, Á.; Torrelo, A. A case of mid-face toddler excoriation syndrome (MiTES). Pediatr. Dermatol. 2020, 37, 345–346. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Singanamalla, B.; Suresh, R.G.; Saini, A.G. Insensitivity to Pain, Self-mutilation, and Neuropathy Associated With PRDM12. Pediatr. Neurol. 2020, 110, 95–96. [Google Scholar] [CrossRef]
- Hasanuddin, S.; Moghe, G.; Reddy, J.S. Hereditary sensory autonomic neuropathy Type VIII: A rare clinical presentation, genomics, diagnosis, and management in an infant. J. Indian Soc. Pedod. Prev. Dent. 2020, 38, 315–318. [Google Scholar] [PubMed]
- Mehmood, S.; Dad, R.; Ahmad, A.; Ullah, M.I.; John, P.; Ali, A.; Hubner, C.A.; Mohyuddin, A.; Hassan, M.J. Structural and functional annotation of PR/SET Domain (PRDM) protein family: In-silico study elaborating role of PRDM12 mutation in congenital insensitivity to pain. Comput. Biol. Chem. 2020, 89, 107382. [Google Scholar] [CrossRef]
- Kokotović, T.; Langeslag, M.; Lenartowicz, E.M.; Manion, J.; Fell, C.W.; Alehabib, E.; Tafakhori, A.; Darvish, H.; Bellefroid, E.J.; Neely, G.G.; et al. PRDM12 Is Transcriptionally Active and Required for Nociceptor Function Throughout Life. Front. Mol. Neurosci. 2021, 14, 720973. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.-A.; Yadav, R.; Gao, D.; Norcliffe-Kaufmann, L.; Slaugenhaupt, S.; Kaufmann, H. Expanding the Genotypic Spectrum of Congenital Sensory and Autonomic Neuropathies Using Whole-Exome Sequencing. Neurol. Genet. 2021, 7, e568. [Google Scholar] [CrossRef] [PubMed]
- Bartesaghi, L.; Wang, Y.; Fontanet, P.; Wanderoy, S.; Berger, F.; Wu, H.; Akkuratova, N.; Bouçanova, F.; Médard, J.-J.; Petitpré, C.; et al. PRDM12 Is Required for Initiation of the Nociceptive Neuron Lineage during Neurogenesis. Cell Rep. 2019, 26, 3484–3492.e4. [Google Scholar] [CrossRef] [Green Version]
- Desiderio, S.; Vermeiren, S.; Van Campenhout, C.; Kricha, S.; Malki, E.; Richts, S.; Fletcher, E.; Vanwelden, T.; Schmidt, B.Z.; Henningfeld, K.A.; et al. Prdm12 Directs Nociceptive Sensory Neuron Development by Regulating the Expression of the NGF Receptor TrkA. Cell Rep. 2019, 26, 3522–3536.e5. [Google Scholar] [CrossRef] [Green Version]
- Landy, M.; Goyal, M.; Casey, K.M.; Liu, C.; Lai, H.C. Loss of Prdm12 during development, but not in mature nociceptors, causes defects in pain sensation. Cell Rep. 2021, 34, 108913. [Google Scholar] [CrossRef]
- Denk, F.; Bennett, D.; McMahon, S.B. Nerve Growth Factor and Pain Mechanisms. Annu. Rev. Neurosci. 2017, 40, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Bataille, A.; Leschiera, R.; L’Hérondelle, K.; Pennec, J.-P.; Le Goux, N.; Mignen, O.; Sakka, M.; Plée-Gautier, E.; Brun, C.; Oddos, T.; et al. In Vitro Differentiation of Human Skin-Derived Cells into Functional Sensory Neurons-Like. Cells 2020, 9, 1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolomietz, E.; Marrano, P.; Yee, K.; Thai, B.; Braude, I.; Chun, K.; Minkin, S.; Kamel-Reid, S.; Minden, M.; Squire, J. Quantitative PCR identifies a minimal deleted region of 120 kb extending from the Philadelphia chromosome ABL translocation breakpoint in chronic myeloid leukemia with poor outcome. Leukemia 2003, 17, 1313–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, A.G.; Nacheva, E.P. A potential role for PRDM12 in the pathogenesis of chronic myeloid leukaemia with derivative chromosome 9 deletion. Leukemia 2003, 18, 178–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huet, S.; Dulucq, S.; Chauveau, A.; Ménard, A.; Chomel, J.-C.; Maisonneuve, H.; Legros, L.; Perrin, M.-C.; Ferrant, E.; Moreilhon, C.; et al. Molecular characterization and follow-up of five CML patients with new BCR-ABL1 fusion transcripts. Genes Chromosom. Cancer 2015, 54, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, L.; Yao, W.; Chen, K.; Xu, H.; Ye, Z. Integrated Analysis of Genetic Abnormalities of the Histone Lysine Methyltransferases in Prostate Cancer. Med. Sci. Monit. 2019, 25, 193–239. [Google Scholar] [CrossRef]
- Reyes, D.A.; Sarría, V.M.S.; Salazar-Viedma, M.; D’Afonseca, V. Histone Methyltransferases Useful in Gastric Cancer Research. Cancer Inform. 2021, 20, 11769351211039862. [Google Scholar] [CrossRef]
- Chen, X.; Wyler, S.C.; Li, L.; Arnold, A.G.; Wan, R.; Jia, L.; Landy, M.A.; Lai, H.C.; Xu, P.; Liu, C. Comparative Transcriptomic Analyses of Developing Melanocortin Neurons Reveal New Regulators for the Anorexigenic Neuron Identity. J. Neurosci. 2020, 40, 3165–3177. [Google Scholar] [CrossRef]
- Jan, S.; Dar, M.I.; Wani, R.; Sandey, J.; Mushtaq, I.; Lateef, S.; Syed, S.H. Targeting EHMT2/ G9a for cancer therapy: Progress and perspective. Eur. J. Pharmacol. 2021, 893, 173827. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rienzo, M.; Di Zazzo, E.; Casamassimi, A.; Gazzerro, P.; Perini, G.; Bifulco, M.; Abbondanza, C. PRDM12 in Health and Diseases. Int. J. Mol. Sci. 2021, 22, 12030. https://doi.org/10.3390/ijms222112030
Rienzo M, Di Zazzo E, Casamassimi A, Gazzerro P, Perini G, Bifulco M, Abbondanza C. PRDM12 in Health and Diseases. International Journal of Molecular Sciences. 2021; 22(21):12030. https://doi.org/10.3390/ijms222112030
Chicago/Turabian StyleRienzo, Monica, Erika Di Zazzo, Amelia Casamassimi, Patrizia Gazzerro, Giovanni Perini, Maurizio Bifulco, and Ciro Abbondanza. 2021. "PRDM12 in Health and Diseases" International Journal of Molecular Sciences 22, no. 21: 12030. https://doi.org/10.3390/ijms222112030
APA StyleRienzo, M., Di Zazzo, E., Casamassimi, A., Gazzerro, P., Perini, G., Bifulco, M., & Abbondanza, C. (2021). PRDM12 in Health and Diseases. International Journal of Molecular Sciences, 22(21), 12030. https://doi.org/10.3390/ijms222112030