The Role of ATRA, Natural Ligand of Retinoic Acid Receptors, on EMT-Related Proteins in Breast Cancer: Minireview
Abstract
:1. Introduction
1.1. Aim of the Study
1.2. Source of the Data
2. Molecular Subtypes of Breast Cancer
- Luminal A (ER+/PR+/HER2−/Ki67−): This is the most common type of breast cancer and tends to be slower-growing and less aggressive than other subtypes. Luminal A tumors are associated with the most favorable prognosis in part because they are usually responsive to hormonal therapy [10]. Tumors also show good differentiation, low grade (1 or 2), and the percentage of their recurrence is low [11]. In addition, the low level of Ki67 protein helps control of cancer growth [8].
- Luminal B (ER+/PR+/HER2− or HER2+/Ki67+): This is a relatively small subgroup of tumors that proliferate significantly more, are less differentiated and express hormone receptors. In addition, this subtype was initially characterized clinically as always being positive for HER2, but more recently has been defined as being highly positive for the protein Ki67 and/or HER2 [12]. Luminal B breast cancers have higher histological than luminal A and recur more often.
- Basal-like (ER−/PR−/HER2−): These cancers are also called triple-negative because they lack these receptors. This subtype, which has the most significant association with women with the BRCA1 and p53 gen mutations, offers the worst prognosis of the other subtypes, in part because treatment advances have lagged behind other molecular subtypes [13]. The majority (about 75%) of triple-negative breast cancers fall into the basal-like subtype defined by gene expression profiling. Proliferative activity is significant. Patients of luminal A and basal subtype form the regional lymph node metastases less frequently [14].
- HER2-enriched (ER−/PR−/HER2+): In the past, this subtype had the worst prognosis; however, the widespread use of targeted therapies for HER2+ cancers have substantially improved outcomes for these patients [15].
- Normal-like (ER+/PR+/HER2−/Ki67−): This subtype has been found to exhibit the genetic characteristics of normal breast samples, although its prognosis is often worse than the luminal A prognosis [8].
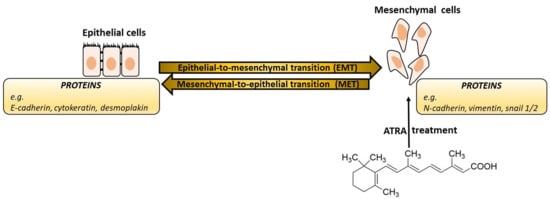
3. Epithelial-Mesenchymal Transition
- EMTs associated with implantation, embryo formation, and organ development are organized to generate different cell types that share common mesenchymal phenotypes. These type 1 EMTs can generate mesenchymal cells that have the potential to subsequently undergo a reverse process—a mesenchymal-epithelial transition (MET) to generate secondary epithelium.
- Type 2 EMTs are associated with tissue regeneration and organ fibrosis. Organic fibrosis, which occurs in many epithelial tissues, is mediated by inflammatory cells and fibroblasts that release various inflammatory signals. Reliable markers for the characterization of mesenchymal products generated by EMT, which occur during the development of fibrosis in various organs, are the following proteins: fibroblast-specific protein 1, a class S100 of the cytoskeletal protein, α-SMA, and collagen I [41,42].
- Type 3 EMTs are associated with cancer progression and metastasis. In the case of this EMT, the cancer cells on the invasive anterior side of the tumors transform into a mesenchymal phenotype. Many in vivo as well as in vitro experiments have shown that cancer cells can acquire a mesenchymal phenotype and express mesenchymal protein markers such as smooth muscle alpha-actin (α-SMA), fibroblast specific protein 1 (FSP1), vimentin, and desmin [43].
4. Natural and Synthetic Retinoid Acid Receptor Ligands and Their Role in EMT
ATRA and Breast Cancer
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANXA1 | Annexin 1 |
| ANXA2 | Annexin 2 |
| APL | Acute promyelocytic leukemia |
| ATRA | All-trans retinoic acid |
| BRD7 | Bromodomain-containing protein 7 |
| CDK1 | Cyclin-dependent kinase 1 |
| 9cRA | 9-cis retinoic acid |
| EMT | Epithelial-mesenchymal transition |
| ER | Estrogen receptor |
| ERBB2 | Receptor tyrosine kinase 2 |
| FGF | Fibroblast growth factors |
| FOXC2 | Forkhead box protein C2 |
| FSP1 | Fibroblast specific protein 1 |
| G3P | Glyceraldehyde 3-phosphate |
| Her2 | Human epidermal growth factor receptor 2 |
| HGF | Hepatocyte growth factor |
| INPP4B | Inositol polyphosphate-4-phosphatase, type II |
| MET | Mesenchymal-epithelial transition |
| MS | Mass spectrometry |
| NB | Neuroblastoma |
| NPM | Nucleophosmin |
| PR | Progesterone receptor |
| RA | Retinoic acid |
| RARs | Retinoic acid receptors |
| RXRs | Retinoid X receptors |
| α-SMA | Smooth muscle alpha-actin |
| TBT-Cl | Tributyltin chloride |
| TGF-β | Transforming growth factor beta |
| TPT-Cl | Triphenyltin chloride |
| VIME | Vimentin |
| YB1 | Y-box-binding protein 1 |
| ZEB | Zinc finger E-box-binding homeobox |
References
- International Agency for Research on Cancer (IARC) in December 2020. Available online: https://www.who.int/news/item/03-02-2021-breast-cancer-now-most-common-form-of-cancer-who-taking-action (accessed on 5 March 2021).
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2016, Section 4: Breast Cancer; National Cancer Institute: Bethesda, MD, USA, 2019; Updated April 2019. Available online: https://seer.cancer.gov/csr/1975_2016/results_merged/sect_04_breast.pdf (accessed on 22 October 2019).
- Qin, X.-J.; Ling, B.X. Proteomic studies in breast cancer. Oncol. Lett. 2012, 3, 735–743. [Google Scholar]
- Tyanova, S.; Albrechtsen, R.; Kronqvist, P.; Cox, J.; Mann, M.; Geiger, T. Proteomic maps of breast cancer subtypes. Nat. Commun. 2016, 7, 10259. [Google Scholar] [CrossRef] [Green Version]
- Yanovich, G.; Agmon, H.; Harel, M.; Sonnenblick, A.; Peretz, T.; Geiger, T. Clinical proteomics of breast cancer reveals a novel layer of breast cancer classification. Cancer Res. 2018, 78, 6001–6010. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.Y.; Qiao, G.A.P.; Liu, Y.Z.; Tian, L.; Hui, N.; Li, J.; Ma, Y.L.; Li, H.; Zhao, Q.Q.; Cao, W.Q.; et al. Overview of all-trans retinoic acid (ATRA) and its analogues: Structures, activities, and mechanisms in acute promyelocytic leukaemia. Eur. J. Med. Chem. 2021, 220, 113451. [Google Scholar] [CrossRef] [PubMed]
- Chlapek, P.; Slavikova, V.; Mazanek, P.; Sterba, J.; Veselska, R. Why differentiation therapy sometimes fails: Molecular mechanisms of resistance to retinoids. Int. J. Mol. Sci. 2018, 19, 132. [Google Scholar] [CrossRef] [Green Version]
- Sasmita, A.O.; Wong, Y.P. Organoids as reliable breast cancer study models: An update. Int. J. Oncol. Res. 2018, 1, 008. [Google Scholar]
- Bouchal, P.; Schubert, O.T.; Faktor, J.; Capkova, L.; Imrichova, H.; Zoufalova, K.; Paralova, V.; Hrstka, R.; Liu, Y.; Ebhardt, H.A.; et al. Breast cancer classification based on proteotypes obtained by SWATH mass spectrometry. Cell Rep. 2019, 28, 832–843. [Google Scholar] [CrossRef]
- Fragomeni, S.M.; Sciallis, A.; Jerus, J.S. Molecular subtypes and local-regional control of breast. Surg. Oncol. Clin. N. Am. 2018, 27, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Abotaleb, M.; Kubatka, P.; Caprnda, M.; Varghese, E.; Zolakova, B.; Zubor, P.; Opatrilova, R.; Kruzliak, P.; Stefanicka, P.; Busselberg, D. Chemotherapeutic agents for the treatment of metastatic breast cancer: An update. Biomed. Pharmacother. 2018, 101, 458–477. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-H.; Hu, P.-H.; Tu, J.-H.; Yu, N.-S. Luminal B breast cancer: Patterns of recurrence and clinical outcome. Oncotarget 2016, 7, 65024–65033. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Yuan, Y.; Liu, G.; Wei, Y. P53 and Ki-67 as prognostic markers in triple-negative breast cancer patients. PLoS ONE 2017, 12, e0172324. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Wang, J.; Xu, B. Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Sig. Transduct. Target Ther. 2019, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- Liskova, A.; Koklesova, L.; Samec, M.; Smejkal, K.; Samuel, S.M.; Varghese, E.; Abotaleb, M.; Biringer, K.; Kudela, E.; Danko, J.; et al. Flavonoids in cancer metastasis. Cancer 2020, 12, 1498. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Swa, H.L.F.; Shaik, A.A.; Lim, L.H.K.; Gunaratne, J. Mass spectrometry based quantitative proteomics and integrative network analysis accentuates modulating roles of annexin-1 in mammary tumorigenesis. Proteomics 2015, 15, 408–418. [Google Scholar] [CrossRef]
- Niu, W.; Luo, Y.; Zhou, Y.; Li, M.; Wu, C.; Duan, Y.; Wang, H.; Fan, S.; Li, Z.; Xiong, W.; et al. BRD7 suppresses invasion and metastasis in breast cancer by negatively regulating YB1-induced EMT. J. Exp. Clin. Cancer Res. 2020, 39, 30. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Wang, Y.; Pan, Q.-Z.; Tang, Y.; Wang, Q.-J.; Pan, K.; Huang, L.-X.; He, J.; Zhao, J.-J.; Jiang, S.-S.; et al. Bromodomain-containing protein 7 (BRD7) as a potential tumor suppressor in hepatocellular carcinoma. Oncotarget 2016, 7, 16248–16261. [Google Scholar] [CrossRef]
- Prieto-García, E.; Díaz-García, C.V.; García-Ruiz, I.; Agulló-Ortuño, M.T. Epithelial-to-mesenchymal transition in tumour progression. Med. Oncol. 2017, 34, 122. [Google Scholar] [CrossRef] [PubMed]
- Morandi, A.; Taddei, M.L.; Chiarugi, P.; Giannoni, E. Targeting the metabolic reprogramming that controls epithelial-to-mesenchymal transition in aggressive tumours. Front. Oncol. 2017, 7, 40. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qin, J.; Liu, Q.; Hong, X.; Li, T.; Zhu, Y.; He, L.; Zheng, B.; Li, M. SNF2H promotes hepatocellular carcinoma proliferation by activating the Wnt/β-catenin signaling pathway. Oncol. Lett. 2016, 12, 1329–1336. [Google Scholar] [CrossRef] [Green Version]
- Strouhalova, D.; Macejova, D.; Lastovickova, M.; Brtko, J.; Bobalova, J. CD44 and vimentin, markers involved with epithelial-mesenchymal transition: A proteomic analysis of sequential proteins extraction of triple-negative breast cancer cells after treatment with all-trans retinoic acid. Gen. Physiol. Biophys. 2020, 39, 399–405. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Wu, H.; Liu, Q.; Pestell, R.G.; Wu, K. The role of CD44 in epithelial–mesenchymal transition and cancer development. OncoTargets Ther. 2015, 8, 3783–3792. [Google Scholar]
- Badaoui, M.; Mimsy-Julienne, C.; Saby, C.; Van Gulick, L.; Peretti, M.; Jeannesson, P.; Morjani, H.; Ouadid-Ahidouch, H. Collagen type 1 promotes survival of human breast cancer cells by overexpressing Kv10.1 potassium and Orai1 calcium channels through DDR1-dependent pathway. Oncotarget 2018, 9, 24653–24671. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Bostan, M.; Caruntu, C.; Ignat, S.R.; Dinescu, S.; Costache, M. Proteomic technology “lens” for epithelial-mesenchymal transition process identification in oncology. Anal. Cell. Pathol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.N.; Xu, H.M. Relationship between collagen IV expression and biological behavior of gastric cancer. World J. Gastroenterol. 2000, 6, 438–439. [Google Scholar] [CrossRef]
- Jung, H.; Kim, B.; Moon, B.I.; Oh, E.-S. Cytokeratin 18 is necessary for initiation of TGF-b1-induced epithelial–mesenchymal transition in breast epithelial cells. Mol. Cell Biochem. 2016, 423, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Yang, L.; Gray, A.; Srivastava, A.K.; Li, C.; Zhang, G.; Cui, T. The role of desmosomes in carcinogenesis. OncoTargets Ther. 2017, 10, 4059–4063. [Google Scholar] [CrossRef] [Green Version]
- Orre, L.M.; Panizza, E.; Kaminskyy, V.O.; Vernet, E.; Graeslund, T.; Zhivotovsky, B.; Lehtioe, J. S100A4 interacts with p53 in the nucleus and promotes p53 degradation. Oncogene 2018, 32, 5531–5540. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Gu, L.-N.; Shan, B.-E.; Geng, C.-Z.; Sang, M.-X. Biomarkers for EMT and MET in breast cancer: An update. Oncol. Lett. 2016, 12, 4869–4876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roussellea, P.; Scoazecb, J.Y. Laminin 332 in cancer: When the extracellular matrix turns signals from cell anchorage to cell. Semin. Cancer Biol. 2020, 62, 149–165. [Google Scholar] [CrossRef]
- Rachow, S.; Zorn-Kruppa, M.; Ohnemus, U.; Kirschner, N.; Vidal-y-Sy, S.; von den Driesch, P.; Bornchen, C.; Eberle, J.; Mildner, M.; Vettorazzi, E.; et al. Occludin is involved in adhesion, apoptosis, differentiation and Ca2+ homeostasis of human keratinocytes: Implications for tumorigenesis. PLoS ONE 2013, 8, e55116. [Google Scholar]
- Kang, E.; Seo, J.; Yoon, H.; Cho, S. The post-translational regulation of epithelial–mesenchymal transition-inducing transcription factors in cancer metastasis. Int. J. Mol. Sci. 2021, 22, 3591. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Gadalla, R.; El-Ghonaimy, E.A.; Samir, O.; Mohamed, H.T.; Hassan, H.; Greve, B.; El-Shinawi, M.; Mohamed, M.M.; Götte, M. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol. Cancer 2017, 16, 57. [Google Scholar] [CrossRef] [Green Version]
- Tailor, D.; Resendez, A.; Garcia-Marques, F.J.; Pandrala, M.; Going, C.C.; Bermudez, A.; Kumar, V.; Rafat, M.; Nambiar, D.K.; Honkala, A.; et al. Y box binding protein 1 inhibition as a targeted therapy for ovarian cancer. Cell Chem. Biol. 2021, 28, 1206–1220.e6. [Google Scholar] [CrossRef] [PubMed]
- Soen, B.; Vandamme, N.; Berx, G.; Schwaller, J.; Van Vlierberghe, P.; Goossens, S. ZEB proteins in leukemia: Friends, foes, or friendly foes? Hemasphere 2018, 2, e43. [Google Scholar] [CrossRef]
- Dekky, B.; Ruff, M.; Bonnier, D.; Legagneux, V.; Théret, N. Proteomic screening identifies the zonula occludens protein ZO-1 as a new partner for ADAM12 in invadopodia-like structures. Oncotarget 2018, 9, 21366–21382. [Google Scholar] [CrossRef] [Green Version]
- Zeisberg, M.; Hanai, J.-I.; Sugimoto, H.; Mammoto, T.; Charytan, D.; Strutz, F.; Kalluri, R. BMP-7 counteracts TGFbeta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat. Med. 2003, 9, 964–968. [Google Scholar] [CrossRef]
- Okada, H.; Danoff, T.M.; Kalluri, R.; Neilson, E.G. Early role of Fsp1 in epithelial-mesenchymal transformation. Am. J. Physiol. 1997, 273, F563–F574. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Weinberg, R.A. Epithelial mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [Green Version]
- Roche, J. The epithelial-to-mesenchymal transition in cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, G.; Rath, B. Mesenchymal-epithelial transition and circulating tumor cells in small cell lung cancer. Adv. Exp. Med. Biol. 2017, 994, 229–245. [Google Scholar]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [Green Version]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Zanetti, A.; Affatato, R.; Centritto, F.; Fratelli, M.; Kurosaki, M.; Barzago, M.M.; Bolis, M.; Terao, M.; Garattini, E.; Paroni, G. All-trans-retinoic acid modulates the plasticity and inhibits the motility of breast cancer cells role of Notch1 and Transforming Growth Factor (Tgf). J. Biol. Chem. 2015, 290, 17690–17709. [Google Scholar] [CrossRef] [Green Version]
- Doi, A.; Ishikawa, K.; Shibata, N.; Ito, E.; Fujimoto, J.; Yamamoto, M.; Shiga, H.; Mochizuki, H.; Kawamura, Y.; Goshima, N.; et al. Enhanced expression of retinoic acid receptor alpha (RARA) induces epithelial-to-mesenchymal transition and disruption of mammary acinar structures. Mol. Oncol. 2015, 9, 355–364. [Google Scholar] [CrossRef]
- Fisher, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Garattini, E.; Gianni, M.; Terao, M. Retinoids as differentiating agents in oncology: A network of interactions with intracellular pathways as the basis for rational therapeutic combinations. Curr. Pharm. Des. 2007, 13, 1375–1400. [Google Scholar] [CrossRef]
- Berbis, P. Retinoids: Mechanisms of action. Ann. Dermatol. Venereol. 2010, 137, S97–S103. [Google Scholar] [CrossRef]
- Brtko, J.; Dvorak, Z. Natural and synthetic retinoid X receptor ligands and their role in selected nuclear receptor action. Biochimie 2020, 179, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Lotan, R. Retinoids and apoptosis: Implication for cancer chemoprevention and therapy. J. Natl. Cancer Inst. 1995, 87, 1655–1657. [Google Scholar] [CrossRef]
- Sun, S.Y.; Lotan, R. Retinoids and their receptors in cancer development and chemoprevention. Crit. Rev. Oncol. Hematol. 2002, 41, 41–55. [Google Scholar] [CrossRef]
- Brtko, J.; Dvorak, Z. Role of retinoids, rexinoids and thyroid hormone in the expression of cytochrome P450 enzymes. Curr. Drug Metab. 2011, 12, 71–88. [Google Scholar] [CrossRef]
- Brtko, J.; Dvorak, Z. Triorganotin compounds—ligands for “rexinoid“ inducible transcription factors: Biological effects. Toxicol. Lett. 2015, 234, 50–58. [Google Scholar] [CrossRef]
- le Maire, A.; Alvarez, S.; Shankaranarayanan, P.; R de Lera, A.; Bourguet, W.; Gronemeyer, H. Retinoid receptors and therapeutic applications of RAR/RXR modulators. Curr. Top. Med. Chem. 2012, 12, 505–527. [Google Scholar] [CrossRef]
- Hunsu, V.O.; Facey, C.O.B.; Fields, J.Z.; Boman, B.M. Retinoids as chemo-preventive and molecular-targeted anticancer therapies. Int. J. Mol. Sci. 2021, 22, 7731. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Giraud, J.; Staedel, C.; Chambonnier, L.; Dubus, P.; Chevret, E.; Bœuf, H.; Gauthereau, X.; Rousseau, B.; Fevre, M.; et al. All-trans retinoic acid targets gastric cancer stem cells and inhibits patient-derived gastric carcinoma tumor growth. Oncogene 2016, 35, 5619–5628. [Google Scholar] [CrossRef]
- Dobrotkova, V.; Chlapek, P.; Mazanek, P.; Sterba, J.; Veselska, R. Traffic lights for retinoids in oncology: Molecular markers of retinoid resistance and sensitivity and their use in the management of cancer differentiation therapy. BMC Cancer 2018, 18, 1059. [Google Scholar] [CrossRef]
- Wille, J.J.; Park, J.Y.; Shealy, Y.F. Cancer chemopreventive retinoids: Validation and analysis of in vivo and in vitro bioassay results. J. Cancer Ther. 2016, 7, 1008–1033. [Google Scholar] [CrossRef] [Green Version]
- Shilkaitis, A.; Green, A.; Christov, K. Retinoids induce cellular senescence in breast cancer cells by RAR-β dependent and independent pathways: Potential clinical implications. Int. J. Oncol. 2015, 47, 35–42. [Google Scholar] [CrossRef]
- Brtko, J. Retinoids, rexinoids and their cognate nuclear receptors: Character and their role in chemoprevention of selected malignant diseases. Biomed. Pap. Med. 2007, 151, 187–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.J.; Gong, M.J.; He, Y.; Li, Q.L.; He, T.C.; Bi, Y. All-trans retinoic acid inhibits proliferation, migration, invasion and induces differentiation of hepa1-6 cells through reversing EMT in vitro. Int. J. Oncol. 2016, 48, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Zhang, H.; Wen, Z.; Gu, Y.M.; Cheng, Y.; Sun, Y.; Zhang, T.T.; Jia, C.W.; Lu, Z.H.; Chen, J. Retinoic acid inhibits pancreatic cancer cell migration and EMT through the downregulation of IL-6 in cancer associated fibroblast cells. Cancer Lett. 2014, 345, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.Y.; Hu, C.Q.; Xu, L.; Cui, J.J.; Tao, L.; Gong, M.J.; Wang, Y.; He, Y.; He, T.C.; Bi, Y. All-trans-retinoic acid inhibits the malignant behaviors of hepatocarcinoma cells by regulating autophagy. Am. J. Transl. Res. 2020, 12, 6793–6810. [Google Scholar] [PubMed]
- Garattini, E.; Bolis, M.; Garattini, S.K.; Fratelli, M.; Centritto, F.; Paroni, G.; Gianni, M.; Zanetti, A.; Pagani, A.; Fisher, J.N.; et al. Retinoids and breast cancer: From basic studies to the clinic and back again. Cancer Treat. Rev. 2014, 40, 739–749. [Google Scholar] [CrossRef]
- Costantini, L.; Molinari, R.; Farinon, B.; Merendino, N. Retinoic acids in the treatment of most lethal solid cancers. J. Clin. Med. 2020, 9, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uray, I.P.; Dmitrovsky, E.; Brown, P.H. Retinoids and rexinoids in cancer prevention: From laboratory to clinic. Semin. Oncol. 2016, 43, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, M.; Leclercq, G. Relevance of breast cancer cell lines as models for breast tumors: An update. Breast Cancer Res. Treat. 2004, 83, 249–289. [Google Scholar] [CrossRef]
- Reinhardt, A.; Liu, H.Y.; Ma, Y.X.; Zhou, Y.G.; Zang, C.B.; Habbel, J.P.; Possinger, K.; Eucker, J. Tumor cell-selective synergism of TRAIL- and ATRA-induced cytotoxicity in breast cancer cells. Anticancer Res. 2018, 38, 2669–2682. [Google Scholar]
- Coyle, K.M.; Dean, C.A.; Thomas, M.L.; Giacomantonio, C.A.; Helyer, L.; Marcato, P. DNA methylation predicts the response of triple-negative breast cancers to all-trans retinoic acid. Cancers 2018, 10, 397. [Google Scholar] [CrossRef] [Green Version]
- Enikeev, A.D.; Komelkov, A.V.; Axelrod, M.E.; Galetsky, S.A.; Kuzmichev, S.A.; Tchevkina, E.M. CRABP1 and CRABP2 protein levels correlate with each other but do not correlate with sensitivity of breast cancer cells to retinoic acid. Biochemistry 2021, 86, 217–229. [Google Scholar] [CrossRef]
- Huang, S.; Chen, Y.; Liang, Z.-M.; Li, N.-N.; Liu, Y.; Zhu, Y.; Liao, D.; Zhou, X.Z.; Lu, K.P.; Yao, Y.; et al. Targeting pin1 by all-trans retinoic acid (ATRA) overcomes tamoxifen resistance in breast cancer via multifactorial mechanisms. Front. Cell Dev. Biol. 2019, 7, 322. [Google Scholar] [CrossRef]
- Kamal, A.H.M.; Han, B.S.; Choi, J.-S.; Cho, K.; Kim, S.Y.; Kim, W.K.; Lee, S.C.; Bae, K.-H. Proteomic analysis of the effect of retinoic acids on the human breast cancer cell line MCF-7. Mol. Biol. Rep. 2014, 41, 3499–3507. [Google Scholar] [CrossRef]
- Flodrova, D.; Benkovska, D.; Macejova, D.; Bialesova, L.; Hunakova, L.; Brtko, J.; Bobalova, J. Proteomic analysis of changes in the protein composition of MCF-7 human breast cancer cells induced by all-trans retinoic acid, 9-cis retinoic acid, and their combination. Toxicol. Lett. 2015, 232, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Croker, A.K.; Allan, A.L. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res. Treat. 2012, 133, 75–87. [Google Scholar] [CrossRef]
- Flodrova, D.; Toporova, L.; Lastovickova, M.; Macejova, D.; Hunakova, L.; Brtko, J.; Bobalova, J. Consequences of the natural retinoid/retinoid X receptor ligands action in human breast cancer MDA-MB-231 cell line: Focus on functional proteomics. Toxicol. Lett. 2017, 281, 26–34. [Google Scholar] [CrossRef]
- Ahrens, T.; Sleeman, J.P.; Schempp, C.M.; Howells, N.; Hofmann, M.; Ponta, H.; Herrlich, P.; Simon, J.C. Soluble CD44 inhibits melanoma tumor growth by blocking cell surface CD44 binding to hyaluronic acid. Oncogene 2001, 20, 3399–3408. [Google Scholar] [CrossRef] [Green Version]
- Li, C.W.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.J.; Asday, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [Green Version]
- Martincuks, A.; Li, P.-C.; Zhao, Q.; Zhang, C.; Li, Y.-J.; Yu, H.; Rodriguez-Rodriguez, L. CD44 in ovarian cancer progression and therapy resistance—A critical role for STAT3. Front. Oncol. 2020, 10, 589601. [Google Scholar] [CrossRef] [PubMed]
- Yaghobi, Z.; Movassaghpour, A.; Talebi, M.; Shadbad, M.A.; Hajiasgharzadeh, K.; Pourvahdani, S.; Baradaran, B. The role of CD44 in cancer chemoresistance: A concise review. Eur. J. Pharmacol. 2021, 903, 174147. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Liu, Y.; Wu, H.; Liu, Q.; Wu, G.S.; Wu, K. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. OncoTargets Ther. 2016, 9, 431–444. [Google Scholar]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [Green Version]
- Eibl, R.H.; Pietsch, T.; Moll, J.; Skroch-Angel, P.; Heider, K.-H.; von Ammon, K.; Wiestler, O.D.; Ponta, H.; Kleihues, P.; Herrlich, P. Expression of variant CD44 epitopes in human astrocytic brain tumors. J. Neurooncol. 1995, 26, 165–170. [Google Scholar] [CrossRef]
- Naor, D.; Nedvetzki, S.; Golan, I.; Melnik, L.; Faitelson, Y. CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 2002, 39, 527–579. [Google Scholar] [CrossRef]
- Ni, X.; Hu, G.; Cai, X. The success and the challenge of all-trans retinoic acid in the treatment of cancer. Crit. Rev. Food Sci. Nutr. 2019, 59, S71–S80. [Google Scholar] [CrossRef]
- Giuli, M.V.; Hanieh, P.N.; Giuliani, E.; Rinaldi, F.; Marianecci, C.; Screpanti, I.; Checquolo, S.; Carafa, M. Current trends in ATRA delivery for cancer therapy. Pharmaceutics 2020, 12, 707. [Google Scholar] [CrossRef]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Medici, N.; Bilancio, A.; Migliaccio, A.; Castoria, G. Breast cancer stem cells: The role of sex steroid receptors. World J. Stem Cells 2019, 11, 594–603. [Google Scholar] [CrossRef]
- Li, N.; Zhu, Y. Targeting liver cancer stem cells for the treatment of hepatocellular carcinoma. Ther. Adv. Gastroenterol. 2019, 12, 1756284818821560. [Google Scholar] [CrossRef]
- Koshiuka, K.; Elstner, E.; Williamson, E.; Said, J.W.; Tada, Y.; Koeffler, H.P. Novel therapeutic approach: Organic arsenical (melarsoprol) alone or with all-trans-retinoic acid markedly inhibit growth of human breast and prostate cancer cells in vitro and in vivo. Br. J. Cancer 2000, 82, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Annuar, S.N.S.; Kamaludin, F.; Awang, N.; Chan, K.M. Cellular basis of organotin(IV) derivatives as anticancer metallodrugs: A review. Front. Chem. 2021, 9, 657599. [Google Scholar] [CrossRef] [PubMed]
- Hunakova, L.; Horvathova, E.; Majerova, K.; Bobal, P.; Otevrel, J.; Brtko, J. Genotoxic effects of tributyltin and triphenyltin isothiocyanates, cognate RXR ligands: Comparison in human breast carcinoma MCF 7 and MDA-MB-231 cells. Int. J. Mol. Sci. 2019, 20, 1198. [Google Scholar] [CrossRef] [Green Version]
- Alama, A.; Tasso, B.; Novelli, F.; Sparatore, F. Organometalic compounds in oncology: Implications of novel organotins as antitumour agents. Drug Discov. Today 2009, 14, 500–508. [Google Scholar] [CrossRef]
- Watanabe, M.; Kakuta, H. Retinoid X receptor antagonists. Int. J. Mol. Sci. 2018, 19, 2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fickova, M.; Macho, L.; Brtko, J. A comparison of the effects of tributyltin chloride and triphenyltin chloride on cell proliferation, proapoptotic p53, Bax, and antiapoptotic Bcl-2 protein levels in human breast cancer MCF-7 cell line. Toxicol. In Vitro 2015, 29, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Strouhalova, D.; Toporova, L.; Lastovickova, M.; Macejova, D.; Bobalova, J.; Brtko, J. Novel insights into the combined effect of triorganotin compounds and all-trans retinoic acid on expression of selected proteins associated with tumor progression in breast cancer cell line MDA-MB-231: Proteomic approach. Gen. Physiol. Biophys. 2019, 38, 135–144. [Google Scholar] [CrossRef]
- Strouhalova, D.; Macejova, D.; Mosna, B.; Bobal, P.; Otevrel, J.; Lastovickova, M.; Brtko, J.; Bobalova, J. Down-regulation of vimentin by triorganotin isothiocyanates-nuclear retinoid X receptor agonists: A proteomic approach. Toxicol. Lett. 2020, 318, 22–29. [Google Scholar] [CrossRef]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jørgensen, C.L.T.; Forsare, C.; Bendahl, P.-O.; Falck, A.-K.; Fernö, M.; Lövgren, K.; Aaltonen, K.; Rydén, L. Expression of epithelial-mesenchymal transition-related markers and phenotypes during breast cancer progression. Breast Cancer Res. Treat. 2020, 181, 369–381. [Google Scholar] [CrossRef]
- Strouhalova, K.; Přechová, M.; Gandalovičová, A.; Brábek, J.; Gregor, M.; Rosel, D. Vimentin intermediate filaments as potential target for cancer treatment. Cancers 2020, 12, 184. [Google Scholar] [CrossRef] [Green Version]




| Protein Name | MW (kDa) | Up/down Regulated during Cancer | Protein Function (www.uniprot.org) | References |
|---|---|---|---|---|
| Annexin 1 (ANX1) | 38.7 | UP |
| [18] |
| Bromodomain-containing protein 7 (BRD7) | 74.1 | Up/tumor suppression |
| [19,20] |
| E-cadherin | 97.5 | Down |
| [21,22] |
| N-cadherin | 99.8 | Up |
| [21,22] |
| β-Catenin | 9.2 | Up |
| [23] |
| CD44 | 81.5 | Up |
| [24,25] |
| Type 1 collagen | 138.9 | Promotes survival of human breast cancer cells by overexpressing Kv10.1 potassium and Orai1 calcium channels. |
| [17,26] |
| Type IV collagen | 164.0 | Down |
| [27,28] |
| Cytokeratin 18 | 48.1 | Down |
| [27,29] |
| Class S100 of cytoskeletal proteins | 9.0–13.0 | Up/Down |
| [17,30] |
| Desmin | 53.5 | Up |
| [17] |
| Desmoplakin | 331.8 | Down |
| [27,31] |
| Fibroblast-specific protein 1 (S100A4) | 11.7 | Up/ overexpressed in a range of different tumor types |
| [17,32] |
| Fibronectin | 2.5 | Up |
| [33] |
| α5 integrin | 114.5 | Up |
| [33] |
| β6 integrin | 85.9 | Up |
| [33] |
| Laminin 1 | 177.6 | Down |
| [27] |
| Laminin 5 | 399.7 | Up |
| [33,34] |
| Mucin 1 | 122.1 | Down |
| [21,22] |
| Occludin | 59.1 | Down |
| [27,35] |
| Smooth muscle alpha-actin (α-SMA) | 42.0 | Up |
| [17,33] |
| Snail | 29.1 | Up |
| [33,36] |
| Syndecan-1 | 32.5 | Up |
| [33,37] |
| Twist | 21 | Up |
| [33,36] |
| Vimentin (VIME) | 53.7 | Up |
| [17,36] |
| Y-box-binding protein 1 | 35.9 | Reduces ovarian cancer cell proliferation |
| [19,38] |
| ZEB proteins ZEB1 ZEB2 | 124.1 133.8 | Up Up |
| [33,36,39] |
| ZO-1 | 187.0 | Down/up |
| [27,35,40] |
| Human Breast Cancer | Lines Description | References |
|---|---|---|
| MCF-10A | no tumorigenic | Reinhardt et al., 2018 [73] |
| BCM-3887 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| BCM-2665 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| BT-20 | ER−, PR−, HER2− | Reinhardt et al., 2018 [73] Coyle et al., 2018 [74] |
| BT-474 | ER+, PR+, HER2+ | Reinhardt et al., 2018 [73] |
| DU4475 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| HBL-100 | epithelial | Enikeev et al., 2021 [75] |
| HCC1187 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| HCC1806 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| HCC1937 | ER−, PR−, HER2− | Coyle et al., 2018 [74] Enikeev et al., 2021 [75] |
| HCC1954 | ER−, PR−, HER2+ | Enikeev et al., 2021 [75] |
| HCC38 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| HCC70 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| MCF-7 | ER+, PR+, HER2− | Reinhardt et al., 2018 [73] Enikeev et al., 2021 [75] Huang et al., 2019 [76] Kamal et al., 2014 [77] Flodrova et al., 2015 [78] |
| MDA-MB-231 | ER−, PR−, HER2− | Strouhalova et al., 2020 [24] Reinhardt et al., 2018 [73] Coyle et al., 2018 [74] Enikeev et al., 2021 [75] Croker and Allan 2012 [79] Flodrova et al., 2017 [80] |
| MDA-MB-453 | ER−, PR−, HER2− | Reinhardt et al., 2018 [73] Coyle et al., 2018 [74] Enikeev et al., 2021 [75] |
| MDA-MB-436 | ER−, PR−, HER2− | Reinhardt et al., 2018 [73] Coyle et al., 2018 [74] |
| MDA-MB-468 | ER−, PR−, HER2− | Coyle et al., 2018 [74] Enikeev et al., 2021 [75] Croker and Allan 2012 [79] |
| SK-BR-3 | ER−, PR−, HER2+ | Reinhardt et al., 2018 [73] Enikeev et al., 2021 [75] |
| SUM-149 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| SUM-159 | ER−, PR−, HER2− | Coyle et al., 2018 [74] |
| T47D | ER+, PR+, HER2− | Reinhardt et al., 2018 [73] Enikeev et al., 2021 [75] Huang et al., 2019 [76] |
| ZR-75-1 | ER+, PR−, HER2− | Reinhardt et al., 2018 [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobal, P.; Lastovickova, M.; Bobalova, J. The Role of ATRA, Natural Ligand of Retinoic Acid Receptors, on EMT-Related Proteins in Breast Cancer: Minireview. Int. J. Mol. Sci. 2021, 22, 13345. https://doi.org/10.3390/ijms222413345
Bobal P, Lastovickova M, Bobalova J. The Role of ATRA, Natural Ligand of Retinoic Acid Receptors, on EMT-Related Proteins in Breast Cancer: Minireview. International Journal of Molecular Sciences. 2021; 22(24):13345. https://doi.org/10.3390/ijms222413345
Chicago/Turabian StyleBobal, Pavel, Marketa Lastovickova, and Janette Bobalova. 2021. "The Role of ATRA, Natural Ligand of Retinoic Acid Receptors, on EMT-Related Proteins in Breast Cancer: Minireview" International Journal of Molecular Sciences 22, no. 24: 13345. https://doi.org/10.3390/ijms222413345
APA StyleBobal, P., Lastovickova, M., & Bobalova, J. (2021). The Role of ATRA, Natural Ligand of Retinoic Acid Receptors, on EMT-Related Proteins in Breast Cancer: Minireview. International Journal of Molecular Sciences, 22(24), 13345. https://doi.org/10.3390/ijms222413345