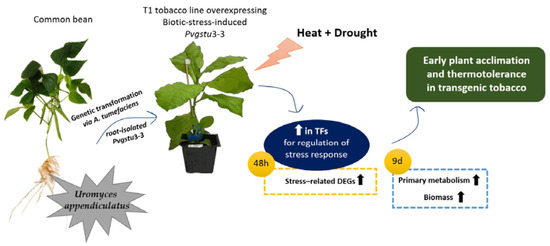
Overexpression of A Biotic Stress-Inducible Pvgstu Gene Activates Early Protective Responses in Tobacco under Combined Heat and Drought
Abstract
:1. Introduction
2. Results
2.1. Evaluation, Relative Expression and Enzymatic Activity of the Transgenes
2.2. Morpho-Physiological Responses
2.3. Effect of Pvgstu3–3 Overexpression on the Transcriptome in Control and Combined Drought and Heat Stress Conditions
2.4. Changes in the Metabolome
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs and Agrobacterium-Mediated Transformation
4.2. Verification of Putative Transgenic Lines, Relative Expression of the 35S-Pvgstu2–2 and -Pvgstu3–3 and Enzymatic Activity
4.3. Application of Abiotic Stress Treatments and Morphophysiological Measurements
4.4. Total RNA Extraction, Library Preparation and Sequencing
Transcriptomics Data Processing and Functional Enrichment Analysis
4.5. Metabolite Extraction and GC–MS Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vile, D.; Pervent, M.; Belluau, M.; Vasseur, F.; Bresson, J.; Muller, B.; Granier, C.; Simonneau, T. Arabidopsis growth under prolonged high temperature and water deficit: Independent or interactive effects? Plant Cell Environ. 2012, 35, 702–718. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Dreesen, F.E.; De Boeck, H.J.; Janssens, I.A.; Nijs, I. Summer heat and drought extremes trigger unexpected changes in productivity of a temperate annual/biannual plant community. Environ. Exp. Bot. 2012, 79, 21–30. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The Combined Effect of Drought Stress and Heat Shock on Gene Expression in Tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [Green Version]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When Defense Pathways Collide. The Response of Arabidopsis to a Combination of Drought and Heat Stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [Green Version]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of Genes Involved in the Response of Arabidopsis to Simultaneous Biotic and Abiotic Stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasch, C.M.; Sonnewald, U. Simultaneous Application of Heat, Drought, and Virus to Arabidopsis Plants Reveals Significant Shifts in Signaling Networks. Plant Physiol. 2013, 162, 1849–1866. [Google Scholar] [CrossRef]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome Responses to Combinations of Stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, P.; Ramegowda, V.; Senthil-Kumar, M. Shared and unique responses of plants to multiple individual stresses and stress combinations: Physiological and molecular mechanisms. Front. Plant Sci. 2015, 6, 723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zandalinas, S.I.; Fichman, Y.; Devireddy, A.R.; Sengupta, S.; Azad, R.K.; Mittler, R. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 13810–13820. [Google Scholar] [CrossRef]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gupta, D.; Nayyar, H. Comparative response of maize and rice genotypes to heat stress: Status of oxidative stress and antioxidants. Acta Physiol. Plant. 2012, 34, 75–86. [Google Scholar] [CrossRef]
- Lo Cicero, L.; Madesis, P.; Tsaftaris, A.; Lo Piero, A.R. Tobacco plants over-expressing the sweet orange tau glutathione transferases (CsGSTUs) acquire tolerance to the diphenyl ether herbicide fluorodifen and to salt and drought stresses. Phytochem. 2015, 116, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xing, X.-J.; Tian, Y.-S.; Peng, R.-H.; Xue, Y.; Zhao, W.; Yao, Q.-H. Transgenic Arabidopsis Plants Expressing Tomato Glutathione S-Transferase Showed Enhanced Resistance to Salt and Drought Stress. PLoS ONE 2015, 10, e0136960. [Google Scholar] [CrossRef] [PubMed]
- Ezaki, B.; Suzuki, M.; Motoda, H.; Kawamura, M.; Nakashima, S. Mechanism of Gene Expression of Arabidopsis in Response to Aluminum Stress. Sci. Technol. 2004, 134, 1672–1682. [Google Scholar]
- Pang, S.; Duan, L.; Liu, Z.; Song, X.; Li, X.; Wang, C. Co-Induction of a Glutathione-S-transferase, a Glutathione Transporter and an ABC Transporter in Maize by Xenobiotics. PLoS ONE 2012, 7, e40712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chronopoulou, E.; Madesis, P.; Tsaftaris, A.; Labrou, N.E. Cloning and Characterization of a Biotic-Stress-Inducible Glutathione Transferase from Phaseolus vulgaris. Appl. Biochem. Biotechnol. 2014, 172, 595–609. [Google Scholar] [CrossRef] [Green Version]
- Ding, N.; Wang, A.; Zhang, X.; Wu, Y.; Wang, R.; Cui, H.; Huang, R.; Luo, Y. Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 2017, 17, 1–15. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate Peroxidase 1 Plays a Key Role in the Response of Arabidopsis thaliana to Stress Combination. J. Biol. Chem. 2008, 283, 34197–34203. [Google Scholar] [CrossRef] [Green Version]
- Demirevska, K.; Simova-Stoilova, L.; Fedina, I.; Georgieva, K.; Kunert, K. Response of Oryzacystatin I Transformed Tobacco Plants to Drought, Heat and Light Stress. J. Agron. Crop. Sci. 2010, 196, 90–99. [Google Scholar] [CrossRef]
- Nahar, K.; Hasanuzzaman, M.; Alam, M.; Rahman, A.; Mahmud, J.-A.; Suzuki, T.; Fujita, M. Insights into spermine-induced combined high temperature and drought tolerance in mung bean: Osmoregulation and roles of antioxidant and glyoxalase system. Protoplasma 2017, 254, 445–460. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Yuan, Z.; Hu, X.; Lu, M.; Wang, W.; Wang, Y. Identification of proteins regulated by ABA in response to combined drought and heat stress in maize roots. Acta Physiol. Plant. 2013, 35, 501–513. [Google Scholar] [CrossRef]
- Kissoudis, C.; Kalloniati, C.; Flemetakis, E.; Madesis, P.; Labrou, N.E.; Tsaftaris, A.; Nianiou-Obeidat, I. Stress-inducible GmGSTU4 shapes transgenic tobacco plants metabolome towards increased salinity tolerance. Acta Physiol. Plant. 2015, 37, 102. [Google Scholar] [CrossRef] [Green Version]
- Broughton, W.J.; Hernández, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.)—Model food legumes. Plant Soil 2003, 252, 55–128. [Google Scholar] [CrossRef] [Green Version]
- Souter, J.R.; Gurusamy, V.; Porch, T.G.; Bett, K.E. Successful Introgression of Abiotic Stress Tolerance from Wild Tepary Bean to Common Bean. Crop Sci. 2017, 57, 1160–1171. [Google Scholar] [CrossRef] [Green Version]
- Mathobo, R.; Marais, D.; Steyn, J.M. The effect of drought stress on yield, leaf gaseous exchange and chlorophyll fluorescence of dry beans (Phaseolus vulgaris L.). Agric. Water Manag. 2017, 180, 118–125. [Google Scholar] [CrossRef] [Green Version]
- Hiz, M.C.; Canher, B.; Niron, H.; Turet, M. Transcriptome Analysis of Salt Tolerant Common Bean (Phaseolus vulgaris L.) under Saline Conditions. PLoS ONE 2014, 9, e92598. [Google Scholar] [CrossRef] [Green Version]
- Dipp, C.C.; Marchese, J.A.; Woyann, L.G.; Bosse, M.A.; Roman, M.H.; Gobatto, D.R.; Paludo, F.; Fedrigo, K.; Kovali, K.K.; Finatto, T. Drought stress tolerance in common bean: What about highly cultivated Brazilian genotypes? Euphytica 2017, 213, 305. [Google Scholar] [CrossRef]
- Chronopoulou, E.; Madesis, P.; Asimakopoulou, B.; Platis, D.; Tsaftaris, A.; Labrou, N.E. Catalytic and structural diversity of the fluazifop-inducible glutathione transferases from Phaseolus vulgaris. Planta 2012, 235, 1253–1269. [Google Scholar] [CrossRef] [Green Version]
- Ganopoulos, I.; Bosmali, I.; Madesis, P.; Tsaftaris, A. Microsatellite genotyping with HRM (High Resolution Melting) analysis for identification of the PGI common bean variety Plake Megalosperma Prespon. Eur. Food Res. Technol. 2012, 234, 501–508. [Google Scholar] [CrossRef]
- Lioi, L.; Piergiovanni, A.R. European Common Bean. In Genetic and Genomic Resources of Grain Legume Improvement, 1st ed.; Singh, M., Upadhyaya, H.D., Bisht, I.S., Eds.; Elsevier Inc.: London, UK, 2013; pp. 11–40. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Zhao, T.-J.; Sun, S.; Liu, Y.; Liu, J.-M.; Liu, Q.; Yan, Y.-B.; Zhou, H.-M. Regulating the Drought-responsive Element (DRE)-mediated Signaling Pathway by Synergic Functions of Trans-active and Trans-inactive DRE Binding Factors in Brassica napus. J. Biol. Chem. 2006, 281, 10752–10759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rucker, K.S.; Kvien, C.K.; Holbrook, C.C.; Hook, J.E. Identification of Peanut Genotypes with Improved Drought Avoidance Traits. Peanut Sci. 1995, 22, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Diao, G.; Wang, Y.; Wang, C.; Yang, C. Cloning and Functional Characterization of a Novel Glutathione S-Transferase Gene from Limonium bicolor. Plant Mol. Biol. Rep. 2011, 29, 77–87. [Google Scholar] [CrossRef]
- Roxas, V.P.; Lodhi, S.A.; Garrett, D.K.; Mahan, J.R.; Allen, R.D. Stress Tolerance in Transgenic Tobacco Seedlings that Overexpress Glutathione S-Transferase/Glutathione Peroxidase. Plant Cell Physiol. 2000, 41, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yu, X.; Ottosen, C.-O.; Rosenqvist, E.; Zhao, L.; Wang, Y.; Yu, W.; Zhao, T.; Wu, Z. Drought stress had a predominant effect over heat stress on three tomato cultivars subjected to combined stress. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanmiya, K.; Suzuki, K.; Egawa, Y.; Shono, M. Mitochondrial small heat-shock protein enhances thermotolerance in tobacco plants. FEBS Lett. 2004, 557, 265–268. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumar, H.; Thamilarasan, S.K.; Shanmugam, A.; Natarajan, S.; Jung, H.-J.; Park, J.-I.; Kim, H.; Chung, M.-Y.; Nou, I.-S. Glutathione Transferases Superfamily: Cold-Inducible Expression of Distinct GST Genes in Brassica oleracea. Int. J. Mol. Sci. 2016, 17, 1211. [Google Scholar] [CrossRef] [Green Version]
- Kayum, A.; Nath, U.K.; Park, J.-I.; Biswas, M.K.; Choi, E.K.; Song, J.-Y.; Kim, H.-T.; Nou, I.-S. Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance. Genes 2018, 9, 84. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Dubey, R.S.; Trivedi, P.K. Expression of a rice Lambda class of glutathione S-transferase, OsGSTL2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J. Hazard. Mater. 2013, 248–249, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Takesawa, T.; Ito, M.; Kanzaki, H.; Kameya, N.; Nakamura, I. Over-expression of ζ glutathione S-transferase in transgenic rice enhances germination and growth at low temperature. Mol. Breed. 2002, 9, 93–101. [Google Scholar] [CrossRef]
- Roxas, V.P.; Smith, R.K.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathioneperoxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Wang, H.; Ran, X.; Li, B.; Zhang, J.; Zhang, H. Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 2016, 14, 1838–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Liu, A.; Yu, J.; Wei, X.; Zhou, R.; Foncéka, D.; Diouf, D.; et al. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wang, Y.; Xin, H.; Li, S.; Liang, Z. Involvement of Ubiquitin-Conjugating Enzyme (E2 Gene Family) in Ripening Process and Response to Cold and Heat Stress of Vitis vinifera. Sci. Rep. 2017, 7, 13290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.-A.; Chang, R.-Z.; Qiu, L.-J. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol. Biol. 2010, 72, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellosillo, T.; Martínez, M.; López, M.A.; Vicente, J.; Cascón, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins Produced by the 9-Lipoxygenase Pathway in Arabidopsis Regulate Lateral Root Development and Defense Responses through a Specific Signaling Cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashi, Y.; Okazaki, Y.; Myouga, F.; Shinozaki, K.; Saito, K. Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 2015, 5, srep10533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Distéfano, A.M.; Valiñas, M.A.; Scuffi, D.; LaMattina, L.; Have, A.T.; García-Mata, C.; Laxalt, A.M. Phospholipase D δ knock-out mutants are tolerant to severe drought stress. Plant Signal. Behav. 2015, 10, e1089371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Labrou, N.E.; Papageorgiou, A.C.; Pavli, O.; Flemetakis, E. Plant GSTome: Structure and functional role in xenome network and plant stress response. Curr. Opin. Biotechnol. 2015, 32, 186–194. [Google Scholar] [CrossRef]
- Yang, G.; Wang, Y.; Xia, D.; Gao, C.; Wang, C.; Yang, C. Overexpression of a GST gene (ThGSTZ1) from Tamarix hispida im-proves drought and salinity tolerance by enhancing the ability to scavenge reactive oxygen species. Plant Cell Tissue Organ Cult. 2014, 117, 99–112. [Google Scholar] [CrossRef]
- Csiszár, J.; Horváth, E.; Váry, Z.; Gallé, Á.; Bela, K.; Brunner, S.; Tari, I. Glutathione transferase supergene family in tomato: Salt stress-regulated expression of representative genes from distinct GST classes in plants primed with salicylic acid. Plant Physiol. Biochem. 2014, 78, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Madesis, P.; Nianiou-Obeidat, I.; Labrou, N.E.; Tsaftaris, A. The Use of Highly Specific GSTs towards the Development of Stress Tolerant Transgenic Plants. In Glutathione: Biochemistry, Mechanisms of Action and Biotechnological Implications; Labrou, N.E., Flemetakis, E., Eds.; Nova Science Pub Inc.: New York, NY, USA, 2013; pp. 263–274. ISBN 9781617617638. [Google Scholar]
- Tian, X.; Wang, Z.; Zhang, Q.; Ci, H.; Wang, P.; Yu, L.; Jia, G. Genome-wide transcriptome analysis of the salt stress tolerance mechanism in Rosa chinensis. PLoS ONE 2018, 13, e0200938. [Google Scholar] [CrossRef]
- George, S.; Venkataraman, G.; Parida, A. A chloroplast-localized and auxin-induced glutathione S-transferase from phreat-ophyte Prosopis juliflora confer drought tolerance on tobacco. J. Plant Physiol. 2010, 167, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liu, Y.; Rao, J.; Wang, G.; Li, H.; Ge, F.; Chen, C. Overexpression of the glutathione S-transferase gene from Pyrus pyrifolia fruit improves tolerance to abiotic stress in transgenic tobacco plants. Mol. Biol. 2013, 47, 515–523. [Google Scholar] [CrossRef]
- Xu, J.; Tian, Y.S.; Xing, X.J.; Peng, R.H.; Zhu, B.; Gao, J.J.; Yao, Q.H. Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol. Plant. 2016, 156, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Sekmen, A.H.; Ozgur, R.; Uzilday, B.; Turkan, I. Reactive oxygen species scavenging capacities of cotton (Gossypium hirsutum) cultivars under combined drought and heat induced oxidative stress. Environ. Exp. Bot. 2014, 99, 141–149. [Google Scholar] [CrossRef]
- Poage, M.; Le Martret, B.; Jansen, M.A.K.; Nugent, G.D.; Dix, P.J. Modification of reactive oxygen species scavenging capacity of chloroplasts through plastid transformation. Plant Mol. Biol. 2011, 76, 371–384. [Google Scholar] [CrossRef]
- Sharma, R.; Sahoo, A.; Devendran, R.; Jain, M. Over-Expression of a Rice Tau Class Glutathione S-Transferase Gene Improves Tolerance to Salinity and Oxidative Stresses in Arabidopsis. PLoS ONE 2014, 9, e92900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dusenge, M.E.; Duarte, A.G.; Way, D.A. Plant carbon metabolism and climate change: Elevated CO2 and temperature impacts on photosynthesis, photorespiration and respiration. New Phytol. 2018, 221, 32–49. [Google Scholar] [CrossRef] [Green Version]
- Rabara, R.C.; Tripathi, P.; Reese, R.N.; Rushton, D.L.; Alexander, D.; Timko, M.P.; Shen, Q.J.; Rushton, P.J. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genom. 2015, 16, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, I.; Sunil, B.; Scheibe, R.; Raghavendra, A.S. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15, 713–722. [Google Scholar] [CrossRef]
- Yao, K.; Wu, Y.Y. Phosphofructokinase and glucose-6-phosphate dehydrogenase in response to drought and bicarbonate stress at transcriptional and functional levels in mulberry. Russ. J. Plant Physiol. 2016, 63, 235–242. [Google Scholar] [CrossRef]
- Trischuk, R.G.; Schilling, B.S.; Wisniewski, M.; Gusta, L.V. Freezing stress: Systems biology to study cold tolerance. In Physiology and Molecular Biology of Stress Tolerance in Plants; Rao, K.V.M., Reddy, K.J., Raghavendra, A.S., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 131–155. [Google Scholar]
- Zhang, X.; Leger, R.J.S.; Fang, W. Pyruvate Accumulation Is the First Line of Cell Defense against Heat Stress in a Fungus. mBio 2017, 8, e01284-17. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, J.; Shao, X.; Li, J. Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 2000, 24, 327–334. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-Responsive Mechanisms in Plant Leaves Revealed by Proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef] [Green Version]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; González, J.A.; Hilal, M.; Prado, F.E. Soluble sugars-metabolism, sensing and abiotic stress a complex network in the life of plants. Plant Signal. Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, R.F.; Irfan, M.; Shakir, H.A.; Khan, M.; Chen, L. Engineering drought tolerance in plants by modification of transcripion and signalling factors. Biotechnol. Biotechnol. Equip. 2020, 34, 781–789. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.-A.; Li, M.-Z.; Wang, S.-M.; Yin, H.-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, R.; Zhou, P.; Ma, X.; Zhou, L.; Wu, Y.; Ullah, Z.; Wang, S. Transcriptome Profiling, Biochemical and Physiological Analyses Provide New Insights towards Drought Tolerance in Nicotiana tabacum L. Genes 2019, 10, 1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Zhu, Z.; Shang, J.; Zhang, S.; Shen, H.; Wu, X.; Zha, D. Transcriptome profiling and gene expression analyses of eggplant (Solanum melongena L.) under heat stress. PLoS ONE 2020, 15, e0236980. [Google Scholar] [CrossRef]
- Wang, L.; Liu, L.; Ma, Y.; Li, S.; Dong, S.; Zu, W. Transcriptome profilling analysis characterized the gene expression patterns responded to combined drought and heat stresses in soybean. Comput. Biol. Chem. 2018, 77, 413–429. [Google Scholar] [CrossRef]
- Pande, A.; Saxena, S.; Thapliyal, M.; Guru, S.K.; Kumar, A.; Arora, S. Role of AP2/EREBP Transcription Factor Family in Environmental Stress Tolerance. Cell Cell. Life Sci. J. 2018, 3, 1–9. [Google Scholar] [CrossRef]
- Dong, C.; Xi, Y.; Chen, X.; Cheng, Z.-M.M. Genome-Wide Identi cation of AP2/EREBP in Fragaria Vesca and Expression Pattern Analysis of The FvDREB Subfamily Under Drought Stress. BMC Plant Biol. 2020. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, T. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genom. 2017, 18, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietz, K.-J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide Identification and Classification of HSF Family in Grape, and Their Transcriptional Analysis under Heat Acclimation and Heat Stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Dai, Y.; He, Y.; Li, C.; Mao, P.; Ma, X. The transcription factors of tall fescue in response to temperature stress. Plant Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Ma, L.; Tian, T.; Lin, R.; Deng, X.W.; Wang, H.; Li, G. Arabidopsis FHY3 and FAR1 Regulate Light-Induced myo-Inositol Bi-osynthesis and Oxidative Stress Responses by Transcriptional Activation of MIPSMol. Plant 2016, 9, 541–557. [Google Scholar]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and devel-opment. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Hu, R.; Gu, T.; Han, J.; Qiu, D.; Su, P.; Feng, J.; Chang, J.; Yang, G.; He, G. Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat. BMC Genom. 2019, 20, 287. [Google Scholar] [CrossRef]
- Xie, Z.-M.; Zou, H.-F.; Lei, G.; Wei, W.; Zhou, Q.-Y.; Niu, C.-F.; Liao, Y.; Tian, A.-G.; Ma, B.; Zhang, W.-K.; et al. Soybean Trihelix Transcription Factors GmGT-2A and GmGT-2B Improve Plant Tolerance to Abiotic Stresses in Transgenic Arabidopsis. PLoS ONE 2009, 4, e6898. [Google Scholar] [CrossRef] [Green Version]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Uribe, L.; O’Connell, M.A. A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J. Exp. Bot. 2006, 57, 1391–1398. [Google Scholar] [CrossRef]
- Polania, J.; Rao, I.M.; Cajiao, C.; Grajales, M.; Rivera, M.; Velasquez, F.; Raatz, B.; Beebe, S.E. Shoot and Root Traits Contribute to Drought Resistance in Recombinant Inbred Lines of MD 23–24 × SEA 5 of Common Bean. Front. Plant Sci. 2017, 8, 296. [Google Scholar] [CrossRef]
- Ramírez-Vallejo, P.; Kelly, J.D. Traits related to drought resistance in common bean. Euphytica 1998, 99, 127–136. [Google Scholar] [CrossRef]
- Teran, J.C.B.M.Y.; Konzen, E.R.; Medina, V.; Palkovic, A.; Ariani, A.; Tsai, S.M.; Gilbert, M.E.; Gepts, P. Root and shoot variation in relation to potential intermittent drought adaptation of Mesoamerican wild common bean (Phaseolus vulgaris L.). Ann. Bot. 2019, 124, 917–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Moschen, S.; Di Rienzo, J.A.; Higgins, J.; Tohge, T.; Watanabe, M.; González, S.; Rivarola, M.; García-García, F.; Dopazo, J.; Hopp, H.E.; et al. Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol. Biol. 2017, 94, 549–564. [Google Scholar] [CrossRef]
- Keunen, E.; Peshev, D.; Vangronsveld, J.; Van Den Ende, W.; Cuypers, A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: Extending the traditional concept. Plant Cell Environ. 2013, 36, 1242–1255. [Google Scholar] [CrossRef]
- Morales, C.; Pino, M.; Del Pozo, A. Phenological and physiological responses to drought stress and subsequent rehydration cycles in two raspberry cultivars. Sci. Hortic. 2013, 162, 234–241. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.S.; Ahmad, D.; Khan, M.A. Utilization of genes encoding osmoprotectants in transgenic plants for enhanced abiotic stress tolerance. Electron. J. Biotechnol. 2015, 18, 257–266. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Jensen, R.G.; Bohnert, H.J. Mannitol Protects against Oxidation by Hydroxyl Radicals. Plant Physiol. 1997, 115, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; Wang, C.; Xiang, B.; Han, R.; Guo, Z. Hydrogen peroxide and nitric oxide mediated cold- and dehydration-induced myo-inositol phosphate synthase that confers multiple resistances to abiotic stresses. Plant Cell Environ. 2013, 36, 288–299. [Google Scholar] [CrossRef]
- Nosarzewski, M.; Downie, A.B.; Wu, B.; Archbold, D.D. The Role of Sorbitol Dehydrogenase in Arabidopsis Thaliana. Funct. Plant Biol. 2012, 39, 462–470. [Google Scholar] [CrossRef]
- Sheveleva, E.V.; Marquez, S.; Chmara, W.; Zegeer, A.; Jensen, R.G.; Bohnert, H.J. Sorbitol-6-phosphate dehydrogenase expression in transgenic tobacco. High amounts of sorbitol lead to necrotic lesions. Plant Physiol. 1998, 117, 831–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kissoudis, C.; Kalloniati, C.; Flemetakis, E.; Madesis, P.; Labrou, N.E.; Tsaftaris, A.; Nianiou-Obeidat, I. Maintenance of met-abolic homeostasis and induction of cytoprotectants and secondary metabolites in alachlor-treated GmGSTU4-overexpressing tobacco plants, as resolved by metabolomics. Plant Biotechnol. Rep. 2015, 9, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Cvikrová, M.; Gemperlová, L.; Martincová, O.; Vanková, R. Effect of drought and combined drought and heat stress on pol-yamine metabolism in proline-over-producing tobacco plants. Plant Physiol. Biochem. 2013, 73, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline Mechanisms of Stress Survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matros, A.; Peshev, D.; Peukert, M.; Mock, H.-P.; Van Den Ende, W. Sugars as hydroxyl radical scavengers: Proof-of-concept by studying the fate of sucralose in Arabidopsis. Plant J. 2015, 82, 822–839. [Google Scholar] [CrossRef]
- Casaretto, J.A.; El-Kereamy, A.; Zeng, B.; Stiegelmeyer, S.M.; Chen, X.; Bi, Y.-M.; Rothstein, S.J. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genom. 2016, 17, 312. [Google Scholar] [CrossRef] [Green Version]
- Axarli, I.A.; Rigden, D.J.; Labrou, N.E. Characterization of the ligandin site of maize glutathione S-transferase I. Biochem. J. 2004, 382, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [Green Version]
- Fichman, Y.; Mittler, R. Rapid systemic signaling during abiotic and biotic stresses: Is the ROS wave master of all trades? Plant J. 2020, 102, 887–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walley, J.W.; Coughlan, S.; Hudson, M.E.; Covington, M.F.; Kaspi, R.; Banu, G.; Harmer, S.L.; Dehesh, K. Mechanical Stress Induces Biotic and Abiotic Stress Responses via a Novel cis -Element. PLOS Genet. 2007, 3, e172. [Google Scholar] [CrossRef] [PubMed]
- Kissoudis, C.; van de Wiel, C.; Visser, R.G.F.; van der Linden, G. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 2014, 5, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative Modifications to Cellular Components in Plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.L.; Shimamoto, K. Sending ROS on a Bullet Train. Sci. Signal. 2009, 2, pe60. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with Stresses: Roles of Calcium- and Calcium/Calmodulin-Regulated Gene Expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [Green Version]
- Walley, J.W.; Dehesh, K. Molecular Mechanisms Regulating Rapid Stress Signaling Networks in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 354–359. [Google Scholar] [CrossRef]
- Benekos, K.; Kissoudis, C.; Nianiou-Obeidat, I.; Labrou, N.E.; Madesis, P.; Kalamaki, M.S.; Makris, A.; Tsaftaris, A.S. Overexpression of a specific soybean GmGSTU4 isoenzyme improves diphenyl ether and chloroacetanilide herbicide tolerance of transgenic tobacco plants. J. Biotechnol. 2010, 150, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Skopelitou, K.; Muleta, A.W.; Pavli, O.; Skaracis, G.N.; Flemetakis, E.; Papageorgiou, A.C.; Labrou, N.E. Overlapping protective roles for glutathione transferase gene family members in chemical and oxidative stress response in Agrobacterium tumefaciens. Funct. Integr. Genom. 2012, 12, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Ricci, G.; Caccuri, A.; Lo Bello, M.; Pastore, A.; Piemonte, F.; Federici, G. Colorimetric and Fluorometric Assays of Glutathione Transferase Based on 7-Chloro-4-nitrobenzo-2-oxa-1,3-diazole. Anal. Biochem. 1994, 218, 463–465. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differ-ential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Michailidis, M.; Karagiannis, E.; Tanou, G.; Karamanoli, K.; Lazaridou, A.; Matsi, T.; Molassiotis, A. Metabolomic and physi-co-chemical approach unravel dynamic regulation of calcium in sweet cherry fruit physiology. Plant Physiol. Biochem. 2017, 116, 68–79. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for statistical and Computing: Vienna, Austria, 2020. [Google Scholar]
- Lawrence, M.A. “ez”: Easy Analysis and Visualization of Factorial Experiments, R package version 4.4-0. CRANpackage repository. 2016.
- Singmann, H.; Bolker, B.; Westfall, J.; Højsgaard, S.; Fox, J.; Lawrence, M.A.; Mertens, U. Afex: Analysis of Factorial Experiments, R package version 0.28-1. CRAN package repository. 2021.
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research, R package version 1.3-3. CRAN package repository. 2020.







| Mid-harvest (Day 9) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Treatment | Shoot Length | THSD | Root Length | THSD | MF | THSD | Number of Leaves | THSD |
| Pvgstu2–2.19 | C | 12 ± 0.85 | a | 9.3 ± 1.4 | a | 6.46 ± 0.41 | a | 11 ± 0.71 | a |
| D | 9.04 ± 0.77 | ab | 7.7 ± 0.62 | ab | 3.48 ± 0.3 | b | 8.6 ± 0.24 | b | |
| T | 9.66 ± 1.09 | ab | 6.9 ± 0.51 | ab | 5.74 ± 0.66 | a | 8.8 ± 0.37 | b | |
| DT | 8.2 ± 0.3 | b | 4.24 ± 0.708 | b | 1.14 ± 0.09 | c | 7.6 ± 0.24 | b | |
| Pvgstu3–3.4 | C | 11.28 ± 0.47 | a | 8.84 ± 0.88 | a | 6.3 ± 0.63 | a | 10 ± 0.55 | a |
| D | 9 ± 0.524 | a | 8.88 ± 0.82 | a | 3.7 ± 0.28 | b | 9 ± 0.32 | a | |
| T | 9.98 ± 0.87 | a | 6.9 ± 1.67 | a | 6.04 ± 0.24 | a | 8.8 ± 0.66 | a | |
| DT | 9 ± 0.464 | a | 5.5 ± 0.55 | a | 1.08 ± 0.14 | c | 6.8 ± 0.37 | b | |
| WT | C | 12.12 ± 1.01 | a | 13.62 ± 2.01 | a | 5.6 ± 0.196 | a | 12.5 ± 1.44 | a |
| D | 10.12 ± 0.96 | a | 8.5 ± 1.67 | ab | 3.45 ± 0.35 | b | 10 ± 0.41 | a | |
| T | 10.87 ± 0.87 | a | 8 ± 1.17 | ab | 4.55 ± 0.3 | a | 9.75 ± 1.25 | a | |
| DT | 9.82 ± 0.64 | a | 6.07 ± 0.47 | b | 0.65 ± 0.08 | c | 5 ± 0 | b | |
| Final Harvest (Day 16) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Treatment | Genotype | Shoot Length | THSD | Root Length | THSD | MF | THSD | Number of Leaves | THSD |
| C | Pvgstu2–2.19 | 15.08 ± 0.64 | a | 9.94 ± 0.77 | a | 8.04 ± 0.5 | ab | 27.45 ± 1.97 | a |
| Pvgstu3–3.4 | 14.24 ± 0.846 | a | 10.23 ± 0.77 | a | 9.28 ± 0.53 | a | 22.33 ± 0.84 | b | |
| WT | 12.02 ± 0.72 | a | 13.2 ± 1.51 | a | 6.8 ± 0.66 | b | 21.75 ± 2.17 | b | |
| D | Pvgstu2–2.19 | 9.89 ± 0.52 | a | 6.88 ± 0.64 | a | 2.46 ± 0.18 | a | 23.1 ± 1.24 | a |
| Pvgstu3–3.4 | 9.37 ± 0.37 | a | 6.01 ± 0.44 | a | 2.5 ± 0.17 | a | 22.7 ± 1.38 | a | |
| WT | 10.85 ± 0.6 | a | 7.07 ± 0.83 | a | 2.17 ± 0.25 | a | 23.75 ± 1.11 | a | |
| T | Pvgstu2–2.19 | 10.5 ± 0.52 | a | 5.09 ± 0.144 | b | 5.6 ± 0.36 | b | 26 ± 1.64 | a |
| Pvgstu3–3.4 | 12.02 ± 0.52 | a | 6.65 ± 0.54 | a | 8.05 ± 0.42 | a | 27.69 ± 1.1 | a | |
| WT | 10.45 ± 0.66 | a | 5.97 ± 0.49 | ab | 5.85 ± 0.36 | b | 24 ± 1.47 | a | |
| DT | Pvgstu2–2.19 | 7.82 ± 0.7 | a | 3.28 ± 0.38 | b | 0.46 ± 0.05 | ab | 15.62 ± 1.33 | a |
| Pvgstu3–3.4 | 9.76 ± 0.58 | a | 5.06 ± 0.82 | ab | 0.64 ± 0.048 | a | 10.43 ± 1.91 | ab | |
| WT | 9.82 ± 0.65 | a | 6.07 ± 0.47 | a | 0.65 ± 0.086 | a | 5 ± 0 | b | |
| Recovery | Pvgstu2–2.19 | 10.42 ± 0.73 | ab | 4.82 ± 0.28 | a | 3.64 ± 0.14 | b | 23.4 ± 1.5 | a |
| Pvgstu3–3.4 | 8.8 ± 0.57 | b | 5.96 ± 0.41 | a | 5.8 ± 0.49 | a | 27.3 ± 3.35 | a | |
| WT | 11.72 ± 0.64 | a | 5.36 ± 0.92 | a | 3.12 ± 0.47 | b | 26.5 ± 2.22 | a | |
| Genotype/Treatment | Pathway Annotation | DEGs | Genes | ||
|---|---|---|---|---|---|
| Total Number (566) | % | Total Number (21,512) | % | ||
| WT vs. Pvgstu3–3.4 under stress-free conditions (C) | Metabolic pathways | 139 | 24.56% | 3944 | 18.33% |
| MAPK signaling pathway | 34 | 6.01% | 768 | 3.57% | |
| NF-kappa B signaling pathway | 31 | 5.48% | 528 | 2.45% | |
| Arachidonic acid metabolism | 14 | 2.47% | 142 | 0.66% | |
| Tryptophan metabolism | 14 | 2.47% | 201 | 0.93% | |
| Peroxisome | 14 | 2.47% | 297 | 1.38% | |
| Retinol metabolism | 12 | 2.12% | 181 | 0.84% | |
| Glycerophospholipid metabolism | 11 | 1.94% | 172 | 0.80% | |
| Linoleic acid metabolism | 10 | 1.77% | 89 | 0.41% | |
| Amino sugar and nucleotide sugar metabolism | 10 | 1.77% | 162 | 0.75% | |
| Glycerolipid metabolism | 9 | 1.59% | 123 | 0.57% | |
| Metabolism of xenobiotics by cytochrome P450 | 9 | 1.59% | 138 | 0.64% | |
| Steroid hormone biosynthesis | 8 | 1.41% | 98 | 0.46% | |
| Phospholipase D signaling pathway | 8 | 1.41% | 131 | 0.61% | |
| Phenylalanine metabolism | 6 | 1.06% | 91 | 0.42% | |
| Total Number (668) | % | Total Number (21,512) | % | ||
| Stress-free WT (C) vs. Pvgstu3–3.4 under 8 h (DT) | Metabolic pathways | 174 | 26.05% | 3944 | 18.33% |
| Starch and sucrose metabolism | 20 | 2.99% | 282 | 1.31% | |
| amino sugar and nucleotide sugar metabolism | 18 | 2.69% | 162 | 0.75% | |
| Galactose metabolism | 14 | 2.10% | 136 | 0.63% | |
| Arachidonic acid metabolism | 14 | 2.10% | 142 | 0.66% | |
| Retinol metabolism | 14 | 2.10% | 181 | 0.84% | |
| Metabolism of xenobiotics by cytochrome P450 | 13 | 1.95% | 138 | 0.64% | |
| Tryptophan metabolism | 13 | 1.95% | 201 | 0.93% | |
| Linoleic acid metabolism | 9 | 1.35% | 89 | 0.41% | |
| Glycine, serine and threonine metabolism | 8 | 1.20% | 91 | 0.42% | |
| Steroid hormone biosynthesis | 8 | 1.20% | 98 | 0.46% | |
| Total Number (1588) | % | Total Number (21,512) | % | ||
| Stress-free WT (C) vs. Pvgstu3–3.4 under 48 h (DT) | Metabolic pathways | 395 | 24.87% | 3944 | 18.33% |
| Carbon metabolism | 55 | 3.46% | 562 | 2.61% | |
| Biosynthesis of amino acids | 40 | 2.52% | 374 | 1.74% | |
| Starch and sucrose metabolism | 39 | 2.46% | 282 | 1.31% | |
| Tryptophan metabolism | 28 | 1.76% | 201 | 0.93% | |
| Arachidonic acid metabolism | 27 | 1.70% | 142 | 0.66% | |
| Amino sugar and nucleotide sugar metabolism | 26 | 1.64% | 162 | 0.75% | |
| Retinol metabolism | 26 | 1.64% | 181 | 0.84% | |
| Metabolism of xenobiotics by cytochrome P450 | 24 | 1.51% | 138 | 0.64% | |
| Glycolysis/gluconeogenesis | 24 | 1.51% | 208 | 0.97% | |
| Glyoxylate and dicarboxylate metabolism | 24 | 1.51% | 224 | 1.04% | |
| Galactose metabolism | 20 | 1.26% | 136 | 0.63% | |
| Folate biosynthesis | 17 | 1.07% | 133 | 0.62% | |
| RNA polymerase | 16 | 1.01% | 116 | 0.54% | |
| Phospholipase D signaling pathway | 16 | 1.01% | 131 | 0.61% | |
| Total Number (3967) | % | Total Number (21,512) | % | ||
| Stress-free Pvgstu3–3.4 (C) vs. Pvgstu3–3.4 at 48 h (DT) | Metabolic pathways | 853 | 21.50% | 3944 | 18.33% |
| Carbon metabolism | 136 | 3.43% | 562 | 2.61% | |
| Endocytosis | 97 | 2.45% | 443 | 2.06% | |
| Biosynthesis of amino acids | 86 | 2.17% | 374 | 1.74% | |
| ABC transporters | 84 | 2.12% | 285 | 1.32% | |
| Apoptosis | 84 | 2.12% | 379 | 1.76% | |
| Peroxisome | 75 | 1.89% | 297 | 1.38% | |
| Pyruvate metabolism | 71 | 1.79% | 311 | 1.45% | |
| Starch and sucrose metabolism | 69 | 1.74% | 282 | 1.31% | |
| Necroptosis | 64 | 1.61% | 226 | 1.05% | |
| Glycolysis/gluconeogenesis | 55 | 1.39% | 208 | 0.97% | |
| Glyoxylate and dicarboxylate metabolism | 55 | 1.39% | 224 | 1.04% | |
| Citrate cycle (TCA cycle) | 55 | 1.39% | 241 | 1.12% | |
| RNA degradation | 53 | 1.34% | 231 | 1.07% | |
| Drug metabolism—other enzymes | 51 | 1.29% | 183 | 0.85% | |
| Base excision repair | 50 | 1.26% | 195 | 0.91% | |
| Tryptophan metabolism | 49 | 1.24% | 201 | 0.93% | |
| Arachidonic acid metabolism | 47 | 1.18% | 142 | 0.66% | |
| Retinol metabolism | 43 | 1.08% | 181 | 0.84% | |
| Metabolism of xenobiotics by cytochrome P450 | 40 | 1.01% | 138 | 0.64% | |
| TFs | WT (C)/Pvgstu3–3.4 (C) | WT (C)/Pvgstu3–3.4 (DT) 48 h | Pvgstu3–3.4 (C)/(DT) 48 h | |||
|---|---|---|---|---|---|---|
| No. of Up- Regulated | No. of Down- Regulated | No. of Up- Regulated | No. of Down- Regulated | No. of Up- Regulated | No. of Down- Regulated | |
| MYB | 27 | 13 | 66 | 33 | 87 | 56 |
| mTERF | - | 2 | 16 | - | 79 | 4 |
| MYB-related | 13 | 12 | 47 | 30 | 70 | 40 |
| FAR1 | 3 | - | 25 | - | 51 | 4 |
| MADS | 1 | 2 | 13 | 5 | 37 | 11 |
| AP2-EREBP | 53 | 49 | 29 | 86 | 32 | 128 |
| NAC | 12 | 7 | 22 | 24 | 31 | 30 |
| Trihelix | - | - | 21 | 2 | 28 | 4 |
| ARF | 1 | 1 | 7 | 6 | 23 | 17 |
| HSF | 3 | 10 | 15 | 13 | 18 | 27 |
| ARR-B | - | - | 2 | - | 13 | - |
| WRKY | 21 | 32 | 12 | 54 | 8 | 71 |
| bZIP | - | 4 | 9 | 6 | 8 | 5 |
| SBP | - | - | 2 | 7 | 8 | 7 |
| Sigma70-like | - | - | 5 | - | 8 | - |
| Pvgstu3–3.4 (DT)/WT (DT) | Pvgstu3–3.4 (C)/WT (C) | ||
|---|---|---|---|
| Metabolites | Log2-Fold Change | Metabolites | Log2-Fold Change |
| Arginine | 20.5 | Glycerol | 2.2 |
| Fructose | 5.3 | Quinic acid | 1.2 |
| Glucose | 4.0 | Ethanolamine | 0.7 |
| Glycine | 3.2 | Sucrose | −0.4 |
| Pantothenate | 3.1 | Fructose | −0.6 |
| Myoinisitol | 2.9 | Glucose | −0.8 |
| Threonic acid | 2.3 | Myoinisitol | −0.8 |
| Tyrosine | 2.2 | Glutamic acid | −0.8 |
| Sorbitol | 2.1 | Nicotine | −0.9 |
| Phenylethanolamine | 1.8 | Malic acid | −0.9 |
| Sucrose | 1.6 | beta_Alanine | −0.9 |
| Xylose | 1.4 | Isoleucine | −0.9 |
| Proline | 1.3 | GABA | −1.0 |
| Threonine | 0.6 | Glycine | −1.0 |
| Arabinose | −0.3 | Arabinose | −1.0 |
| Meso_eryrthitol | −0.3 | Proline | −1.0 |
| Glycerol | −0.5 | Sorbitol | −1.0 |
| Phenylalanine | −0.5 | Glyceric acid | −1.0 |
| Ethanolamine | −0.5 | Valine | −1.0 |
| Serine | −0.6 | Phenylalanine | −1.0 |
| Glutamine | −0.7 | Threose | −1.0 |
| Alanine | −0.9 | Oxoproline | −1.0 |
| beta_Alanine | −0.9 | Mannitol | −1.0 |
| Lysine | −0.9 | ||
| GABA | −0.9 | ||
| Galactose | −1.0 | ||
| Tryptophan | −1.0 | ||
| Genotype | Treatment | Analysis group |
|---|---|---|
| WT | Control; 0 h | Group 1 |
| Pvgstu3–3.4 | Control; 0 h | Group 2 |
| DT combined stress; 8 h | Group 3 | |
| DT combined stress; 48 h | Group 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stavridou, E.; Voulgari, G.; Michailidis, M.; Kostas, S.; Chronopoulou, E.G.; Labrou, N.E.; Madesis, P.; Nianiou-Obeidat, I. Overexpression of A Biotic Stress-Inducible Pvgstu Gene Activates Early Protective Responses in Tobacco under Combined Heat and Drought. Int. J. Mol. Sci. 2021, 22, 2352. https://doi.org/10.3390/ijms22052352
Stavridou E, Voulgari G, Michailidis M, Kostas S, Chronopoulou EG, Labrou NE, Madesis P, Nianiou-Obeidat I. Overexpression of A Biotic Stress-Inducible Pvgstu Gene Activates Early Protective Responses in Tobacco under Combined Heat and Drought. International Journal of Molecular Sciences. 2021; 22(5):2352. https://doi.org/10.3390/ijms22052352
Chicago/Turabian StyleStavridou, Evangelia, Georgia Voulgari, Michail Michailidis, Stefanos Kostas, Evangelia G. Chronopoulou, Nikolaos E. Labrou, Panagiotis Madesis, and Irini Nianiou-Obeidat. 2021. "Overexpression of A Biotic Stress-Inducible Pvgstu Gene Activates Early Protective Responses in Tobacco under Combined Heat and Drought" International Journal of Molecular Sciences 22, no. 5: 2352. https://doi.org/10.3390/ijms22052352
APA StyleStavridou, E., Voulgari, G., Michailidis, M., Kostas, S., Chronopoulou, E. G., Labrou, N. E., Madesis, P., & Nianiou-Obeidat, I. (2021). Overexpression of A Biotic Stress-Inducible Pvgstu Gene Activates Early Protective Responses in Tobacco under Combined Heat and Drought. International Journal of Molecular Sciences, 22(5), 2352. https://doi.org/10.3390/ijms22052352