Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation
Abstract
:1. Introduction
2. Results
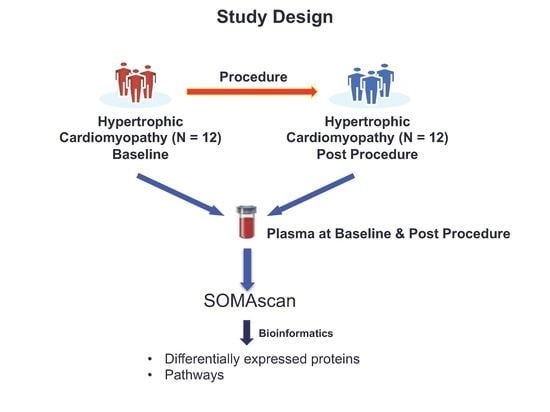
2.1. Patient Cohort Characteristics
2.2. SOMAscan Plasma Proteomics Demonstrates Within-Person Stability of Distinct Protein Fingerprints
2.3. SOMAscan Enables the Detection of Myectomy-Related Protein Expression and Identification of Differentially Expressed Proteins in Plasma That Distinguish between the Preoperative and Postoperative State
2.4. Ingenuity Pathway Analysis Reveals Protein Interaction Networks, Upstream Regulators, and Biological Processes Relevant to HCM
3. Discussion
4. Materials and Methods
4.1. Study Patients
4.2. Blood Sample Processing
4.3. SOMAscan Proteomics Profiling
4.4. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LVOT | Left Ventricular Outflow Tract |
| HCM | Hypertrophic Cardiomyopathy |
| PCA | Principal Component Analysis |
| ECM | Extracellular Matrix |
| TNF | Tumor Necrosis Factor |
| IFNγ | Interferon γ |
| TGFβ1 | Transforming Growth Factor β1 |
| POSTN | Periostin |
| NAMPT | Nicotinamide Phosphoribosyl-transferase |
| EGF | Epidermal Growth Factor |
| VEGF | Vascular Endothelial Growth Factor |
References
- Hathout, Y.; Brody, E.; Clemens, P.R.; Cripe, L.; DeLisle, R.K.; Furlong, P.; Gordish-Dressman, H.; Hache, L.; Henricson, E.; Hoffman, E.P.; et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 2015, 112, 7153–7158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, D.; Sinha, S.; Shen, D.; Kuhn, E.W.; Keyes, M.J.; Shi, X.; Benson, M.D.; O’Sullivan, J.F.; Keshishian, H.; Farrell, L.A.; et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 2016, 134, 270–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, Y.J.; Hasegawa, K.; Kochav, S.M.; Mohajer, P.; Jung, J.; Maurer, M.S.; Reilly, M.P.; Fifer, M.A. Application of proteomics profiling for biomarker discovery in hypertrophic cardiomyopathy. J. Cardiovasc. Transl. Res. 2019. [Google Scholar] [CrossRef]
- Fang, L.; Ellims, A.H.; Beale, A.L.; Taylor, A.J.; Murphy, A.; Dart, A.M. Systemic inflammation is associated with myocardial fibrosis, diastolic dysfunction, and cardiac hypertrophy in patients with hypertrophic cardiomyopathy. Am. J. Transl. Res. 2017, 9, 5063–5073. [Google Scholar] [PubMed]
- Landry, N.M.; Cohen, S.; Dixon, I.M.C. Periostin in cardiovascular disease and development: A tale of two distinct roles. Basic Res. Cardiol. 2018, 113, 1. [Google Scholar] [CrossRef]
- Hu, W.; Wei, R.; Wang, L.; Lu, J.; Liu, H.; Zhang, W. Correlations of MMP-1, MMP-3, and MMP-12 with the degree of atherosclerosis, plaque stability and cardiovascular and cerebrovascular events. Exp. Ther. Med. 2018, 15, 1994–1998. [Google Scholar] [CrossRef]
- Liu, S.L.; Bajpai, A.; Hawthorne, E.A.; Bae, Y.; Castagnino, P.; Monslow, J.; Pure, E.; Spiller, K.L.; Assoian, R.K. Cardiovascular protection in females linked to estrogen-dependent inhibition of arterial stiffening and macrophage MMP12. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Jeong, M.H.; Kim, H.J.; Pyun, J.H.; Choi, K.S.; Lee, D.I.; Solhjoo, S.; O’Rourke, B.; Tomaselli, G.F.; Jeong, D.S.; Cho, H.; et al. Cdon deficiency causes cardiac remodeling through hyperactivation of WNT/beta-catenin signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E1345–E1354. [Google Scholar] [CrossRef] [Green Version]
- Chapouly, C.; Hollier, P.L.; Guimbal, S.; Cornuault, L.; Gadeau, A.P.; Renault, M.A. Desert hedgehog-driven endothelium integrity is enhanced by gas1 (growth arrest-specific 1) but negatively regulated by Cdon (cell adhesion molecule-related/downregulated by oncogenes). Arterioscler. Thromb Vasc. Biol. 2020, 40, e336–e349. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.; Oka, S.I.; Imai, N.; Huang, C.Y.; Ralda, G.; Zhai, P.; Ikeda, Y.; Ikeda, S.; Sadoshima, J. Both gain and loss of Nampt function promote pressure overload-induced heart failure. Am. J. Physiol Heart Circ. Physiol. 2019, 317, H711–H725. [Google Scholar] [CrossRef]
- Travelli, C.; Colombo, G.; Mola, S.; Genazzani, A.A.; Porta, C. NAMPT: A pleiotropic modulator of monocytes and macrophages. Pharmacol. Res. 2018, 135, 25–36. [Google Scholar] [CrossRef]
- Zlatanova, I.; Pinto, C.; Bonnin, P.; Mathieu, J.R.R.; Bakker, W.; Vilar, J.; Lemitre, M.; Voehringer, D.; Vaulont, S.; Peyssonnaux, C.; et al. Iron regulator hepcidin impairs macrophage-dependent cardiac repair after injury. Circulation 2019, 139, 1530–1547. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Gabrielsen, A.; Agardh, H.E.; Wan, M.; Wetterholm, A.; Wong, C.H.; Hedin, U.; Swedenborg, J.; Hansson, G.K.; Samuelsson, B.; et al. Expression of 5-lipoxygenase and leukotriene A4 hydrolase in human atherosclerotic lesions correlates with symptoms of plaque instability. Proc. Natl. Acad. Sci. USA 2006, 103, 8161–8166. [Google Scholar] [CrossRef] [Green Version]
- Kuusisto, J.; Karja, V.; Sipola, P.; Kholova, I.; Peuhkurinen, K.; Jaaskelainen, P.; Naukkarinen, A.; Yla-Herttuala, S.; Punnonen, K.; Laakso, M. Low-grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart 2012, 98, 1007–1013. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Weng, Z.; Xia, Q.; Cao, C.; Leak, R.K.; Han, L.; Xiao, J.; Graham, S.H.; Cao, G. Intracerebroventricular delivery of recombinant NAMPT deters inflammation and protects against cerebral ischemia. Transl Stroke Res. 2019, 10, 719–728. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, R.; Wang, H.; Tao, Q.; Lin, X.; Ge, S.; Zhai, Z. Nicotinamide phosphate transferase (NAMPT) increases in plasma in patients with acute coronary syndromes, and promotes macrophages to M2 polarization. Int. Heart J. 2018, 59, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Gogiraju, R.; Bochenek, M.L.; Schafer, K. Angiogenic endothelial cell signaling in cardiac hypertrophy and heart failure. Front. Cardiovasc. Med. 2019, 6, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cruijsen, H.; Giaccone, G.; Hoekman, K. Epidermal growth factor receptor and angiogenesis: Opportunities for combined anticancer strategies. Int. J. Cancer 2005, 117, 883–888. [Google Scholar] [CrossRef]
- Mendez-Barbero, N.; Gutierrez-Munoz, C.; Blazquez-Serra, R.; Martin-Ventura, J.L.; Blanco-Colio, L.M. Tumor necrosis factor-like weak inducer of apoptosis (TWEAK)/fibroblast growth factor-inducible 14 (Fn14) axis in cardiovascular diseases: Progress and challenges. Cells 2020, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.F.; Li, J.; Bertrand, A.; Casbon, A.J.; Lin, J.H.; Maltseva, I.; Werb, Z. Protective effects of matrix metalloproteinase-12 following corneal injury. J. Cell Sci. 2013, 126, 3948–3960. [Google Scholar] [CrossRef] [Green Version]
- Marconcini, L.; Marchio, S.; Morbidelli, L.; Cartocci, E.; Albini, A.; Ziche, M.; Bussolino, F.; Oliviero, S. C-fos-induced growth factor/vascular endothelial growth factor D induces angiogenesis in vivo and in vitro. Proc. Natl. Acad. Sci. USA 1999, 96, 9671–9676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, E.R.; Berg, K.L.; Einstein, D.B.; Lee, P.S.; Pixley, F.J.; Wang, Y.; Yeung, Y.G. Biology and action of colony—Stimulating factor-1. Mol. Reprod. Dev. 1997, 46, 4–10. [Google Scholar] [CrossRef]
- Esensten, J.H.; Helou, Y.A.; Chopra, G.; Weiss, A.; Bluestone, J.A. CD28 costimulation: From mechanism to therapy. Immunity 2016, 44, 973–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguado, B.A.; Schuetze, K.B.; Grim, J.C.; Walker, C.J.; Cox, A.C.; Ceccato, T.L.; Tan, A.C.; Sucharov, C.C.; Leinwand, L.A.; Taylor, M.R.G.; et al. Transcatheter aortic valve replacements alter circulating serum factors to mediate myofibroblast deactivation. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [Green Version]
- Kraemer, S.; Vaught, J.D.; Bock, C.; Gold, L.; Katilius, E.; Keeney, T.R.; Kim, N.; Saccomano, N.A.; Wilcox, S.K.; Zichi, D.; et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: A SOMAmer-based, streamlined multiplex proteomic assay. PLoS ONE 2011, 6, e26332. [Google Scholar] [CrossRef] [Green Version]
- Fong, T.G.; Chan, N.Y.; Dillon, S.T.; Zhou, W.; Tripp, B.; Ngo, L.H.; Otu, H.H.; Inouye, S.K.; Vasunilashorn, S.M.; Cooper, Z.; et al. Identification of plasma proteome signatures associated with surgery using SOMAscan. Ann. Surg. 2019. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]









| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Demographics | ||||||||||||
| age at myectomy | 37 | 43 | 55 | 56 | 73 | 73 | 58 | 63 | 52 | 76 | 63 | 73 |
| female | yes | no | yes | yes | yes | yes | no | no | yes | yes | yes | yes |
| nyha class ≥ 3 | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | no |
| Med Hx | ||||||||||||
| Prior AF | no | no | no | no | no | no | no | yes | no | no | yes | no |
| Prior VT/VF | yes | no | no | no | yes | no | no | no | no | no | no | no |
| Prior NS VT | no | no | no | no | no | no | no | no | no | no | ||
| Prior syncope | yes | no | no | no | no | no | no | no | yes | no | no | no |
| Fam Hx SCD | yes | no | no | no | no | no | no | no | no | no | no | no |
| Fam Hx HCM | yes | no | yes | no | no | no | no | no | no | yes | no | no |
| Comorbidities | pituitary adenoma | none | CAD, HTN, HLD, COPD, DM2, OSA, Spinal Stenosis | CAD, HTN, HLD | CAD, pituitary adenoma, DI, HLD | OSA, HTN, CAD, HLD | HTN, HLD, CAD | HLD | HTN, cerebral aneurysm, diverticulosis | AS, HLD, pulmonary HTN, hypothyroidism | HTN, HLD, OSA, morbid obesity, GERD | HTN, HLD, AS, MR, GERD |
| Meds | ||||||||||||
| beta blocker | yes | yes | yes | yes | yes | no | yes | yes | yes | yes | yes | yes |
| calcium channel blocker | no | no | no | yes | no | yes | no | no | no | no | yes | no |
| ACE or ARB | no | no | no | no | no | no | no | no | yes | no | no | no |
| Diuretic Use | no | no | no | yes | no | no | yes | no | no | no | yes | no |
| loop diuretic | no | no | no | no | no | no | yes | no | no | no | yes | no |
| thiazide | no | no | no | yes | no | no | no | no | no | no | no | no |
| potassium sparing | no | no | no | no | no | no | no | no | no | no | no | no |
| disopyramide | no | no | no | no | no | no | no | no | no | no | no | no |
| amiodarone | no | no | no | no | no | no | no | no | no | no | no | no |
| Physiological measurements | ||||||||||||
| LA size (mm) | NF | 52 | 46 | 43 | 45 | 49 | 54 | 57 | 43 | 50 | 47 | 45 |
| systolic blood pressure | 135 | 128 | 110 | 170 | 105 | 126 | 142 | 126 | 148 | 128 | 122 | 117 |
| diastolic blood pressure | 80 | 82 | 80 | 70 | 56 | 78 | 90 | 78 | 74 | 78 | 70 | 70 |
| IVS thickness (mm) | 23 | 13 | 15 | 15 | 17 | 15 | 15 | 18 | 14 | 23 | 17 | 18 |
| Posterior wall thickness | NF | 12 | 12 | 9.4 | 13 | 8.9 | 8.7 | 14 | 12 | 13 | 11 | 11 |
| LVEF (%) | 65 | 70 | 65 | 65 | 70 | 65 | 65–70 | 65 | 65 | 60 | 65 | 65 |
| LVEDD (mm) | NF | 45 | 36 | 46 | 31 | 31 | 48 | 44 | 42 | 34 | 38 | 41 |
| LVESD (m) | NV | 29 | 23 | 27 | 20 | 22 | 33 | 25 | 26 | 23 | 29 | 29 |
| SAM | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| MR | mild | mod | mild | mod | mod-severe | mild | trace | trace | trace | mod | trace | mod |
| LVOT gradient rest (mm Hg) | 0 | 60 | 35 | 90 | 100 | 0 | 0 | 100 | 0 | 150 | 100 | 55 |
| LVOT gradient provocation (mm Hg) | 60 | 85 | 150 | 110 | 145 | 100 | ||||||
| LVOT gradient postop visit | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LGE on MRI | ND (ICD) | none | mild | none | ND (ICD) | none | none | none | none | mild | transmural septal consistent with prior ablation | mild |
| Surgical Procedure | extended septal myectomy with release of papillary muscle attachment | extended septal myectomy with mitral valve repair | extended septal myectomy, CABGx1 | extended septal myectomy, mitral valve repair, right hemithorax drainage (SC injury) | extended myectomy, cabgx1 | extended septal myectomy | septal myectomy and MV repair | extended septal myectomy, MAZE | extended septal myectomy | extended septal myectomy, aortic valve replacement | extended septal myectomy, MAZE | extended septal myectomy, aortic valve replacement |
| Pathogenic HCM Variant | MYBPC3 | NF | NF | NF | MYBPC3 | NF | ND | ND | ND | NF | ND | ND |
| Up-Regulated and Down-Regulated Proteins between Hypertrophic Cardiomyopathy Patients before (PRE) and after Surgical Myectomy (POST) | |||||||
|---|---|---|---|---|---|---|---|
| Increased in POST as compared to PRE | |||||||
| SomaId | TargetFullName | Target | Gene Symbol | p-Value | BH adj. p-Value | Mean FC | Median FC |
| SL000522 | Macrophage metalloelastase | MMP-12 | MMP12 | 4.6376E-05 | 0.06052076 | 1.31 | 1.68 |
| SL008574 | Osteomodulin | OMD | OMD | 0.00066949 | 0.14561424 | 1.37 | 1.39 |
| SL005084 | Periostin | Periostin | POSTN | 8.6775E-05 | 0.05662100 | 1.34 | 1.36 |
| SL000468 | Immunoglobulin M | IgM | IGHM IGJ IGK@ IGL@ | 0.0014402 | 0.23493316 | 1.22 | 1.35 |
| SL003167 | C-X-C motif chemokine 13 | BLC | CXCL13 | 0.02038433 | 0.64881839 | 1.57 | 1.33 |
| SL007471 | Collectin-12 | COLEC12 | COLEC12 | 0.02738612 | 0.68728638 | 1.14 | 1.32 |
| SL000339 | Carbonic anhydrase 2 | carbonic anhydrase II | CA2 | 0.02469152 | 0.67130076 | 1.17 | 1.28 |
| SL008059 | 40S ribosomal protein S3 | RS3 | RPS3 | 0.01568655 | 0.60208673 | 1.17 | 1.26 |
| SL006406 | Plexin-C1 | PLXC1 | PLXNC1 | 0.00306231 | 0.33302632 | 1.18 | 1.22 |
| SL006108 | CD5 antigen-like | CD5L | CD5L | 0.00986478 | 0.51494130 | 1.16 | 1.20 |
| SL007429 | Transmembrane glycoprotein NMB | GPNMB | GPNMB | 0.01371202 | 0.55919313 | 1.12 | 1.19 |
| SL008360 | Leucine-rich repeat transmembrane protein FLRT2 | FLRT2 | FLRT2 | 0.00469783 | 0.47159018 | 1.17 | 1.19 |
| SL000542 | Integrin alpha-IIb: beta-3 complex | gpIIbIIIa | ITGA2B ITGB3 | 0.02127866 | 0.66115832 | 1.24 | 1.17 |
| SL005185 | Interleukin-23 receptor | IL-23 R | IL23R | 0.04845637 | 0.80045010 | 1.10 | 1.17 |
| SL013969 | Kynureninase | KYNU | KYNU | 0.00272817 | 0.32366037 | 1.25 | 1.15 |
| SL003993 | Bone morphogenetic protein 6 | BMP-6 | BMP6 | 0.00927918 | 0.55042406 | 1.12 | 1.14 |
| SL008609 | Low affinity immunoglobulin gamma Fc region receptor III-B | FCG3B | FCGR3B | 0.04122137 | 0.76848410 | 1.08 | 1.14 |
| SL002508 | Interleukin-18-binding protein | IL-18 BPa | IL18BP | 0.044693 | 0.78816716 | 1.11 | 1.13 |
| SL005764 | Scavenger receptor cysteine-rich type 1 protein M130 | sCD163 | CD163 | 0.0091015 | 0.62512959 | 1.13 | 1.13 |
| SL012740 | A disintegrin and metalloproteinase with thrombospondin motifs 15 | ATS15 | ADAMTS15 | 0.04123163 | 0.75784896 | 1.12 | 1.13 |
| SL011532 | Persulfide dioxygenase ETHE1, mitochondrial | ETHE1 | ETHE1 | 0.04060736 | 0.76800881 | 1.16 | 1.12 |
| SL014092 | Cell adhesion molecule-related/down-regulated by oncogenes | CDON | CDON | 0.00474673 | 0.44246350 | 1.19 | 1.12 |
| SL000455 | Transcription factor AP-1 | c-Jun | JUN | 0.04687702 | 0.79447425 | 1.06 | 1.10 |
| SL007179 | Ephrin type-B receptor 2 | EPHB2 | EPHB2 | 0.01035377 | 0.51967950 | 1.14 | 1.09 |
| SL014896 | Ankyrin-2 | ANK2 | ANK2 | 0.00753706 | 0.65572461 | 1.15 | 1.09 |
| SL010379 | Semaphorin-3A | Semaphorin 3A | SEMA3A | 0.01190177 | 0.53557973 | 1.23 | 1.08 |
| SL010613 | Interleukin-17 receptor D | IL-17 RD | IL17RD | 0.0085244 | 0.69527115 | 1.06 | 1.04 |
| SL007206 | Thrombospondin-2 | TSP2 | THBS2 | 0.00906359 | 0.65711062 | 1.22 | 1.03 |
| SL005159 | Erythropoietin receptor | EPO-R | EPOR | 0.04481069 | 0.77970596 | 1.23 | 1.03 |
| Decreased in POST as compared to PRE | |||||||
| SL004536 | Hepcidin | LEAP-1 | HAMP | 0.00022828 | 0.09930021 | −2.78 | −3.13 |
| SL000420 | Ferritin | Ferritin | FTH1 FTL | 0.03198654 | 0.09930021 | −1.85 | −2.04 |
| SL004820 | Phosphoglycerate mutase 1 | Phosphoglycerate mutase 1 | PGAM1 | 0.0167906 | 0.09930021 | −1.82 | −1.52 |
| SL007049 | Cystatin-F | CYTF | CST7 | 0.00966243 | 0.09930021 | −1.69 | −1.45 |
| SL007100 | Leukotriene A-4 hydrolase | LKHA4 | LTA4H | 0.0002564 | 0.09930021 | −1.36 | −1.41 |
| SL003310 | Vascular endothelial growth factor A, isoform 121 | VEGF121 | VEGFA | 0.02172054 | 0.09930021 | −1.29 | −1.40 |
| SL008835 | Asialoglycoprotein receptor 1 | ASGR1 | ASGR1 | 0.02304106 | 0.09930021 | −1.42 | −1.35 |
| SL002602 | Trefoil factor 2 | Trefoil factor 2 | TFF2 | 0.02012063 | 0.09930021 | −1.22 | −1.31 |
| SL000546 | Prolactin | PRL | PRL | 0.01198619 | 0.09930021 | −1.41 | −1.30 |
| SL006830 | Complement factor H-related protein 5 | complement factor H-related 5 | CFHR5 | 0.01622241 | 0.09930021 | −1.15 | −1.27 |
| SL000586 | Thrombin | Thrombin | F2 | 0.01871806 | 0.09930021 | −1.52 | −1.26 |
| SL004351 | Interleukin-25 | IL-17E | IL25 | 0.02671673 | 0.09930021 | −1.12 | −1.24 |
| SL007953 | Pyridoxal kinase | PDXK | PDXK | 0.0397993 | 0.09930021 | −1.27 | −1.23 |
| SL003685 | Nicotinamide phosphoribosyltransferase | PBEF | NAMPT | 0.00239742 | 0.09930021 | −1.23 | −1.22 |
| SL004805 | Cell adhesion molecule 1 | Nectin-like protein 2 | CADM1 | 0.03249705 | 0.09930021 | −1.18 | −1.21 |
| SL004064 | Phospholipase A2 | GIB | PLA2G1B | 0.03789695 | 0.09930021 | −1.35 | −1.21 |
| SL004814 | Coactosin-like protein | Coactosin-like protein | COTL1 | 0.03793821 | 0.09930021 | −1.18 | −1.19 |
| SL004365 | Tumor necrosis factor ligand superfamily member 12 | TWEAK | TNFSF12 | 0.03165744 | 0.09930021 | −1.24 | −1.18 |
| SL003915 | Kallikrein-8 | kallikrein 8 | KLK8 | 0.02436652 | 0.09930021 | −1.17 | −1.18 |
| SL000603 | Trypsin-1 | Trypsin | PRSS1 | 0.00905271 | 0.09930021 | −1.15 | −1.17 |
| SL003643 | Glutathione S-transferase P | Glutathione S-transferase Pi | GSTP1 | 0.02132777 | 0.09930021 | −1.18 | −1.16 |
| SL004859 | Tumor necrosis factor receptor superfamily member 18 | GITR | TNFRSF18 | 0.03458126 | 0.09930021 | −1.26 | −1.15 |
| SL000164 | Myoglobin | Myoglobin | MB | 0.00944586 | 0.09930021 | −1.18 | −1.15 |
| SL009207 | Dynactin subunit 2 | Dynactin subunit 2 | DCTN2 | 0.00153051 | 0.09930021 | −1.17 | −1.14 |
| SL004147 | Interleukin-10 receptor subunit beta | IL-10 Rb | IL10RB | 0.03733233 | 0.09930021 | −1.09 | −1.13 |
| SL014983 | Histone H2B type 2-E | H2B2E | HIST2H2BE | 0.04030205 | 0.09930021 | −1.25 | −1.12 |
| SL005361 | Apolipoprotein D | Apo D | APOD | 0.0223943 | 0.09930021 | −1.10 | −1.12 |
| SL010619 | Tryptase gamma | TPSG1 | TPSG1 | 0.0091463 | 0.09930021 | −1.14 | −1.12 |
| SL006378 | S-formylglutathione hydrolase | Esterase D | ESD | 0.03428547 | 0.09930021 | −1.20 | −1.11 |
| SL005575 | Galactoside 3(4)-L-fucosyltransferase | Fucosyltransferase 3 | FUT3 | 0.00112912 | 0.09930021 | −1.10 | −1.11 |
| SL000314 | Complement C3b | C3b | C3 | 0.02655588 | 0.09930021 | −1.45 | −1.11 |
| SL004304 | Syntaxin-1A | STX1a | STX1A | 0.00039638 | 0.09930021 | −1.10 | −1.10 |
| SL016129 | Protein FAM107B | FAM107B | FAM107B | 0.03490093 | 0.09930021 | −1.22 | −1.10 |
| SL004366 | Tumor necrosis factor receptor superfamily member 12A | TWEAKR | TNFRSF12A | 0.01632066 | 0.09930021 | −1.12 | −1.09 |
| SL004118 | Tartrate-resistant acid phosphatase type 5 | TrATPase | ACP5 | 0.01179264 | 0.09930021 | −1.07 | −1.08 |
| SL010376 | Membrane metallo-endopeptidase-like 1 | MMEL2 | MMEL1 | 0.01299823 | 0.09930021 | −1.06 | −1.08 |
| SL000587 | Thyroglobulin | Thyroglobulin | TG | 0.03721233 | 0.09930021 | −1.08 | −1.08 |
| SL014093 | Ectonucleoside triphosphate diphosphohydrolase 3 | ENTP3 | ENTPD3 | 0.04419097 | 0.09930021 | −1.07 | −1.07 |
| SL002662 | Coagulation Factor XI | Coagulation Factor XI | F11 | 0.00927585 | 0.09930021 | −1.07 | −1.07 |
| SL000084 | Epidermal growth factor | EGF | EGF | 0.03368098 | 0.09930021 | −1.14 | −1.07 |
| SL004742 | Afamin | Afamin | AFM | 0.03493132 | 0.09930021 | −1.07 | −1.07 |
| SL004724 | Histone acetyltransferase KAT6A | MOZ | KAT6A | 0.02857323 | 0.09930021 | −1.06 | −1.07 |
| SL008865 | Proteasome subunit alpha type-2 | PSA2 | PSMA2 | 0.0275171 | 0.09930021 | −1.11 | −1.06 |
| SL010469 | Hemojuvelin | RGM-C | HFE2 | 0.04126795 | 0.09930021 | −1.08 | −1.06 |
| SL003919 | Kallikrein-14 | kallikrein 14 | KLK14 | 0.0465157 | 0.09930021 | −1.07 | −1.06 |
| SL003774 | Bcl-2-like protein 2 | Apoptosis regulator Bcl-W | BCL2L2 | 0.04714896 | 0.09930021 | −1.11 | −1.05 |
| SL004637 | Macrophage-stimulating protein receptor | MSP R | MST1R | 0.01888127 | 0.09930021 | −1.10 | −1.05 |
| SL006992 | Matrilin-3 | MATN3 | MATN3 | 0.01065689 | 0.09930021 | −1.07 | −1.04 |
| SL007673 | Netrin-4 | NET4 | NTN4 | 0.01398477 | 0.09930021 | −1.12 | −1.04 |
| SL004648 | Tumor necrosis factor ligand superfamily member 14 | LIGHT | TNFSF14 | 0.02543942 | 0.09930021 | −1.10 | −1.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larson, A.; Libermann, T.A.; Bowditch, H.; Das, G.; Diakos, N.; Huggins, G.S.; Rastegar, H.; Chen, F.Y.; Rowin, E.J.; Maron, M.S.; et al. Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation. Int. J. Mol. Sci. 2021, 22, 2474. https://doi.org/10.3390/ijms22052474
Larson A, Libermann TA, Bowditch H, Das G, Diakos N, Huggins GS, Rastegar H, Chen FY, Rowin EJ, Maron MS, et al. Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation. International Journal of Molecular Sciences. 2021; 22(5):2474. https://doi.org/10.3390/ijms22052474
Chicago/Turabian StyleLarson, Amy, Towia A. Libermann, Heather Bowditch, Gaurav Das, Nikolaos Diakos, Gordon S. Huggins, Hassan Rastegar, Frederick Y. Chen, Ethan J. Rowin, Martin S. Maron, and et al. 2021. "Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation" International Journal of Molecular Sciences 22, no. 5: 2474. https://doi.org/10.3390/ijms22052474
APA StyleLarson, A., Libermann, T. A., Bowditch, H., Das, G., Diakos, N., Huggins, G. S., Rastegar, H., Chen, F. Y., Rowin, E. J., Maron, M. S., & Chin, M. T. (2021). Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation. International Journal of Molecular Sciences, 22(5), 2474. https://doi.org/10.3390/ijms22052474