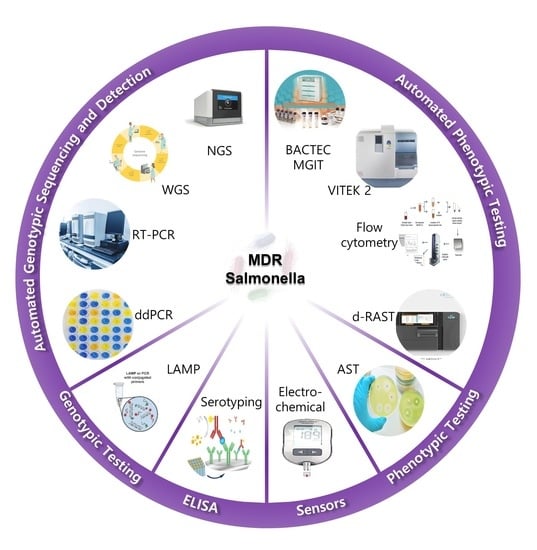
Recent Advances in the Detection of Antibiotic and Multi-Drug Resistant Salmonella: An Update
Abstract
:1. Introduction
2. Conventional Isolation, Enrichment and Detection Methods
2.1. Rapid Salmonella Detection Methods
2.1.1. Enzyme-Linked Immuno-Sorbent Assay (ELISA)
2.1.2. Nucleic Acid Assay Techniques
3. Automated Whole Genome Sequencing
4. Automated Phenotypic Testing
4.1. Manual and Semi-Automated Antimicrobial Susceptibility Test (AST)
4.2. Detection of Intracellular Resistance Salmonella Using Flow Cytometry
5. Emerging Biosensors
5.1. Optical Biosensors
5.2. Electrochemical Biosensors
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistant |
| AST | Antimicrobial susceptibility test |
| BGA | Brilliant green agar |
| BSA | Bismuth-sulfite agar |
| CNT | Carbon nanotubes |
| CDC | Center for Disease Control and Prevention |
| CRISPR-MVLST | CRISPR-multi-locus virulence sequence typing |
| CV | Cyclic voltammetry |
| DD | Digital droplet |
| DPV | Differential pulse voltammetry |
| EIS | Electrochemical impedance spectroscopy |
| ESBLs | Extended-spectrum β-lactamases |
| EU-CAST | European Committee on Antimicrobial Susceptibility Testing |
| FC | Flow Cytometry |
| FDA | Federal Drug Agency |
| FISH | Fluorescence in situ hybridization |
| GNP | Gold nanoparticulate |
| HE | Hektoen enteric |
| HP | Hydrogen peroxide |
| MDR | Multi-Drug Resistant |
| ME | Major errors |
| MIC | Minimum inhibitory concentration |
| MLST | Multi-locus sequence typing |
| MLVA | Multiple-locus variable number of tandem repeats analysis |
| NGS | Next-generation sequencing |
| NTS | Non-typhoidal Salmonella |
| PCR | Polymerase Chain Reaction |
| ROS | Reactive oxygen species |
| RT-PCR | Real-time PCR |
| RV | Rappaport-Vassiliadis |
| SERS | Surface Enhanced Raman Spectroscopy |
| SNP | Single-nucleotide polymorphism |
| SS | Salmonella-Shigella |
| SPR | Surface plasmon resonance |
| TMD | Transition metal dichalcogenide |
| TSI | Triple sugar iron |
| VBNC | Viable but nonculturable |
| VME | Very major errors |
| WGS | Whole genome sequencing |
| XLD | Xylose-lysine-deoxycholate agar |
References
- Grimont, P.A.; Weill, F.X. Antigenic. Formulae of Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella: Geneva, Switzerland, 2007. [Google Scholar]
- Bugarel, M.; Granier, S.A.; Weill, F.X.; Fach, P.; Brisabois, A. A multiplex real-time PCR assay targeting virulence and resistance genes in Salmonella enterica serotype Typhimurium. BMC Microbiol. 2011, 11, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Lee, K.M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control. 2015, 47, 264–276. [Google Scholar] [CrossRef]
- Stephen Inbaraj, B.; Chen, B.H. Nanomaterial-based sensors for detection of foodborne bacterial pathogens and toxins as well as pork adulteration in meat products. J. Food. Drug. Anal. 2016, 24, 15–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooq, U.; Yang, Q.; Ullah, M.W.; Wang, S. Bacterial biosensing: Recent advances in phage-based bioassays and biosensors. Biosens. Bioelectron. 2018, 118, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.; Lee, S.; Yang, Y.A.; Song, J. The role of typhoid toxin in Salmonella Typhi virulence. Yale J. Biol. Med. 2017, 90, 283–290. [Google Scholar]
- Tamamura, Y.; Tanaka, K.; Uchida, I. Characterization of pertussis-like toxin from Salmonella spp. that catalyzes ADP-ribosylation of G proteins. Sci. Rep. 2017, 7, 2653. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.A.; Betteken, M.I.; Guo, X.; Altier, C.; Duhamel, G.E.; Wiedmann, M. The typhoid toxin produced by the Non-typhoidal Salmonella enterica Serotype javiana is required for induction of a DNA damage response in Vitro and systemic spread in Vivo. MBio 2018, 9, e00467-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, R.A.; Wiedmann, M. The ADP-ribosylating toxins of Salmonella. Toxins 2019, 11, 416. [Google Scholar] [CrossRef] [Green Version]
- Outbreaks of Salmonella Infections Linked to Backyard Poultry. Final Update; 2019. Available online: https://www.cdc.gov/salmonella/backyardpoultry-05-19/index.html/ (accessed on 2 March 2020).
- CDC 2011. National Enteric Disease Surveillance: Salmonella Annual Summary; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2011.
- Gebreyes, W.A.; Thakur, S. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob. Agents. Chemother. 2005, 49, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, A. Antibiotic stewardship: Why we must, how we can. Clevel. Clin. J. Med. 2017, 84, 673–679. [Google Scholar] [CrossRef]
- USDA; FSIS; Food and Drug Administration (FDA). Retail Meat Interim Report. 2014–2015. Available online: https://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM498134.pdf (accessed on 26 November 2020).
- Selvin, J.; Maity, D.; Sajayan, A.; Kiran, G.S. Revealing antibiotic resistance in therapeutic and dietary probiotic supplements. J. Glob. Antimicrob. Resist. 2020, 22, 202–205. [Google Scholar] [CrossRef]
- Xiang, Y.; Li, F.; Dong, N.; Tian, S.; Zhang, H.; Du, X.; Zhou, X.; Xu, X.; Yang, H.; Xie, J.; et al. Investigation of a Salmonellosis Outbreak Caused by Multidrug Resistant Salmonella Typhimurium in China. Front. Microbiol. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, H.; Tang, Y.; Li, Q.; Jiao, X. A Multidrug-resistant Monophasic Salmonella bla CTX-M-14 in a Transferable IncHI2 Plasmid from a Healthy Catering Worker in China. Infect. Drug. Resist. 2020, 13, 3569–3574. [Google Scholar] [CrossRef] [PubMed]
- Besser, J.M. Salmonella epidemiology: A whirlwind of change. Food Microbiol. 2018, 71, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Aydindogan, E.; Guler Celik, E.; Timur, S. Paper-Based Analytical Methods for Smartphone Sensing with Functional Nanoparticles: Bridges from Smart Surfaces to Global Health. Anal. Chem. 2018, 90, 12325–12333. [Google Scholar] [CrossRef]
- Kanchi, S.; Sabela, M.I.; Mdluli, P.S.; Inamuddin; Bisetty, K. Smartphone based bioanalytical and diagnosis applications: A review. Biosens. Bioelectron. 2018, 102, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Domesle, K.J.; Ge, B. Loop-Mediated Isothermal Amplification for Salmonella Detection in Food and Feed: Current Applications and Future Directions. Foodborne Pathog. Dis. 2018, 15, 309–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.V.T.; Thomas, J.V.; Dewi, G.; Johnson, T.; Noll, S.; Cardona, C.; Kollanoor-Johny, A. Effects of Multiple Alternatives-To-Antibiotic Interventions on Multidrug-Resistant Salmonella Heidelberg in Turkey Poults. In Proceedings of the 2017 PSA Annual Meeting, Orlando, FL, USA, 17–20 July 2017; Volume 96, p. 24. [Google Scholar]
- Carrique-Mas, J.J.; Davies, R.H. Sampling and bacteriological detection of Salmonella in poultry and poultry premises: A review. Rev. Sci. Tech. 2008, 27, 665–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anvarinejad, M.; Pouladfar, G.R.; Pourabbas, B.; Shahidi, M.A.; Rafaatpour, N.; Dehyadegari, M.A.; Abbasi, P.; Mardaneh, J. Detection of Salmonella spp. with the BACTEC 9240 automated blood culture system in 2008–2014 in Southern Iran (Shiraz): Biogrouping, MIC, and antimicrobial susceptibility profiles of isolates. Jundishapur J. Microbiol. 2016, 9, e26505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behera, B.; Vishnu, G.A.; Chatterjee, S.; Sreekumar, N.; Nagabhushan, A.; Rajendran, N.; Prathik, B.; Pandya, H.J. Emerging technologies for antibiotic susceptibility testing. Biosens. Bioelectron. 2019, 142, 111552. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Huang, Y.; Duan, M.; Xing, K.; You, X.; Zhou, H.; Liu, Y.; Liu, C.; Liu, D.; Lai, W. Biosensing multiplexer based on immunochromatographic assay for rapid and high-throughput classification of Salmonella serogroups. Sens. Actuators B Chem. 2019, 282, 317–321. [Google Scholar] [CrossRef]
- Hulme, J. Recent advances in the detection of methicillin resistant Staphylococcus aureus (MRSA). Biochip J. 2017, 11, 89–100. [Google Scholar] [CrossRef]
- Spanò, S.; Ugalde, J.E.; Galán, J.E. Delivery of a Salmonella Typhi Exotoxin from a Host Intracellular Compartment. Cell Host Microbe 2008, 3, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, H.; Mutharasan, R. Review of biosensors for foodborne pathogens and toxins. Sens. Actuators B Chem. 2013, 183, 535–549. [Google Scholar] [CrossRef]
- Valderrama, W.B.; Dudley, E.G.; Doores, S.; Cutter, C.N. Commercially available rapid methods for detection of selected food-borne pathogens. Crit. Rev. Food Sci. Nutr. 2016, 56, 1519–1531. [Google Scholar] [CrossRef]
- Singh, G.; Sithebe, A.; Enitan, A.M.; Kumari, S.; Bux, F.; Stenström, T.A. Comparison of droplet digital PCR and quantitative PCR for the detection of Salmonella and its application for river sediments. J. Water Health. 2017, 15, 505–508. [Google Scholar] [CrossRef]
- Bugarel, M.; Tudor, A.; Lonergan, G.H.; Nightingale, K.K. Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J. Microbiol. Methods 2017, 134, 14–20. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, J.; Zhang, B.; Sun, S.; Chang, W. Characterization of integrons and resistance genes in Salmonella isolates from farm animals in Shandong province, China. Front. Microbiol. 2017, 8, 1300. [Google Scholar] [CrossRef]
- Chiu, C.H.; Su, L.H.; Chu, C.H.; Wang, M.H.; Yeh, C.M.; Weill, F.X.; Chu, C. Detection of multidrug-resistant Salmonella enterica serovar typhimurium phage types DT102, DT104, and U302 by multiplex PCR. J. Clin. Microbiol. 2006, 44, 2354–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Trinetta, V.; Shi, X.; Noll, L.W.; Magossi, G.; Zheng, W.; Porter, E.P.; Cernicchiaro, N.; Renter, D.G.; Nagaraja, T.G. A multiplex real-time PCR assay, based on invA and pagC genes, for the detection and quantification of Salmonella enterica from cattle lymph nodes. J. Microbiol. Methods 2018, 148, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Mustapha, A. Multiplex TaqMan® detection of pathogenic and multidrug resistant Salmonella. Int. J. Food Microbiol. 2013, 166, 213–218. [Google Scholar] [CrossRef]
- Trinh, K.T.L.; Trinh, T.N.D.; Lee, N.Y. Fully integrated and slidable paper-embedded plastic microdevice for point-of-care testing of multiple foodborne pathogens. Biosens. Bioelectron. 2019, 135, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.N.D.; Lee, N.Y. A foldable isothermal amplification microdevice for fuchsin-based colorimetric detection of multiple foodborne pathogens. Lab Chip 2019, 19, 1397–1405. [Google Scholar] [CrossRef]
- Papadakis, G.; Pantazis, A.K.; Ntogka, M.; Parasyris, K.; Theodosi, G.I.; Kaprou, G.; Gizeli, E. 3D-printed Point-of-Care Platform for Genetic Testing of Infectious Diseases Directly in Human Samples Using Acoustic Sensors and a Smartphone. ACS Sens. 2019, 4, 1329–1336. [Google Scholar] [CrossRef]
- Ricke, S.C.; Kim, S.A.; Shi, Z.; Park, S.H. Molecular-based identification and detection of Salmonella in food production systems: Current perspectives. J. Appl. Microbiol. 2018, 125, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Shah, D.H.; Casavant, C.; Hawley, Q.; Addwebi, T.; Call, D.R.; Guard, J. Salmonella Enteritidis strains from poultry exhibit differential responses to acid stress, oxidative stress, and survival in the egg albumin. Foodborne Pathog. Dis. 9, 258–264. [CrossRef] [Green Version]
- Brown, E.; Dessai, U.; Mcgarry, S.; Gerner-Smidt, P. Use of Whole-Genome Sequencing for Food Safety and Public Health in the United States. Foodborne Pathog. Dis. 2019, 16, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.K.; Sharma, P.; McMillan, E.A.; Jackson, C.R.; Hiott, L.M.; Woodley, T.; Humayoun, S.B.; Barrett, J.B.; Frye, J.G.; McClelland, M. Genomic comparison of diverse Salmonella serovars isolated from swine. PLoS ONE 2019, 14, e0224518. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.; Seribelli, A.A.; Cazentini Medeiros, M.I.; Rodrigues, D.d.P.; de MelloVarani, A.; Luo, Y.; Allard, M.W.; Falcão, J.P. Phylogenetic and antimicrobial resistance gene analysis of Salmonella Typhimurium strains isolated in Brazil by whole genome sequencing. PLoS ONE 2018, 13, e0201882. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Migura, L.; Sunde, M.; Karlsmose, S.; Veldman, K.; Schroeter, A.; Guerra, B.; Granier, S.A.; Perrin-Guyomard, A.; Gicquel-Bruneau, M.; Franco, A.; et al. Establishing streptomycin epidemiological cut-off values for Salmonella and Escherichia coli. Microb. Drug Resist. 2012, 18. [Google Scholar] [CrossRef] [Green Version]
- McDermott, P.F.; Tyson, G.H.; Kabera, C.; Chen, Y.; Li, C.; Folster, J.P.; Ayers, S.L.; Lam, C.; Tate, H.P.; Zhao, S. Whole-genome sequencing for detecting antimicrobial resistance in non-typhoidal Salmonella. Antimicrob. Agents Chemother. 2016, 60, 5515–5520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, L.M.; Wiedmann, M.; den Bakker, H.; Siler, J.; Warchocki, S.; Kent, D.; Lyalina, S.; Davis, M.; Sischo, W.; Besser, T.; et al. Whole-Genome Sequencing of Drug-Resistant Salmonella enterica Isolates from Dairy Cattle and Humans in New York and Washington States Reveals Source and Geographic Associations. Appl. Environ. Microbiol. 2017, 83, e00140-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mensah, N.; Tang, Y.; Cawthraw, S.; Abuoun, M.; Fenner, J.; Thomson, N.R.; Mather, A.E.; Petrovska-Holmes, L. Determining antimicrobial susceptibility in Salmonella enterica serovar Typhimurium through whole genome sequencing: A comparison against multiple phenotypic susceptibility testing methods. BMC Microbiol. 2019, 19, 148. [Google Scholar] [CrossRef] [PubMed]
- Zwe, Y.H.; Chin, S.F.; Kohli, G.S.; Aung, K.T.; Yang, L.; Yuk, H.G. Whole genome sequencing(WGS) fails to detect antimicrobial resistance (AMR) from heteroresistant subpopulation of Salmonella enterica. Food Microbiol. 2020, 91, 103530. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, J.; le Hello, S.; Doublet, B.; Granier, S.A.; Hendriksen, R.S.; Florian Fricke, W.; Ceyssens, P.J.; Gomart, C.; Billman-Jacobe, H.; Holt, K.E.; et al. Global phylogenomics of multidrug-resistant salmonella enterica serotype kentucky ST198. Microb. Genom. 2019, 5, e000269. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. MIC and Zone Diameter Distributions and ECOFFs. EUCAST, 2018. Available online: http://www.eucast.org/mic_distributions_and_ecoffs/ (accessed on 24 December 2020).
- Choi, J.R.; Yong, K.W.; Choi, J.Y.; Cowie, A.C. Emerging point-of-care technologies for food safety analysis. Sensors 2019, 19, 817. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Masum, F.; Jeon, J.S. Recent Developments of Chip-based Phenotypic Antibiotic Susceptibility Testing. Biochip J. 2019. [Google Scholar] [CrossRef]
- Hu, Q.; Yu, Y.; Gu, D.; Xie, L.; Chen, X.; Xu, N.; Ruan, J.J.; Dowson, C.; Ruan, B.H. Detection of “hidden” Antimicrobial Drug Resistance. ACS Infect. Dis. 2019, 5, 1252–1263. [Google Scholar] [CrossRef]
- Garner, O.; Traczewski, M.M.; Beasley, D.; Harrington, A.; DesJarlais, S.; Hastey, C.; Brookman, R.; Lockett, Z.; Chau, J. 667. Multicenter Assessment of Enterobacterales, Salmonella spp. and Pseudomonas aeruginosa Using Updated CLSI Levofloxacin Breakpoints on MicroScan Dried Gram-Negative MIC Panels. Open Forum Infect. Dis. 2020, 7 (Suppl. S1), S388–S389. [Google Scholar] [CrossRef]
- Leonard, H.; Colodner, R.; Halachmi, S.; Segal, E. Recent Advances in the Race to Design a Rapid Diagnostic Test for Antimicrobial Resistance. ACS Sens. 2018, 3, 2202–2217. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Ambriz-Aviña, V.; Contreras-Garduño, J.A.; Pedraza-Reyes, M. Applications of Flow Cytometry to Characterize Bacterial Physiological Responses. BioMed Res. Int. 2014, 461941. [Google Scholar] [CrossRef] [PubMed]
- Van Meervenne, E.; van Coillie, E.; Kerckhof, F.M.; Devlieghere, F.; Herman, L.; de Gelder, L.S.P.; Top, E.M.; Boon, N. Strain-specific transfer of antibiotic resistance from an environmental plasmid to foodborne pathogens. J. Biomed. Biotechnol. 2012, 834598. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.H.; Tzeng, Y.L.; Dickson, R.M. FAST: Rapid determinations of antibiotic susceptibility phenotypes using label-free cytometry. Cytom. Part A. 2018, 93, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Rishi, P.; Bhagat, N.R.; Thakur, R.; Pathania, P. Tackling Salmonella Persister Cells by Antibiotic-Nisin Combination via Mannitol. Indian J. Microbiol. 2018, 58, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Claudi, B.; Spröte, P.; Chirkova, A.; Personnic, N.; Zankl, J.; Schürmann, N.; Schmidt, A.; Bumann, D. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 2014, 158, 722–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vohra, P.; Vrettou, C.; Hope, J.C.; Hopkins, J.; Stevens, M.P. Nature and consequences of Interactions between Salmonella enterica serovar Dublin and host cells in cattle. Vet. Res. 2019, 50, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsolis, R.M.; Adams, L.G.; Ficht, T.A.; Baumler, A.J. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 1999, 67, 4879–4885. [Google Scholar] [CrossRef] [Green Version]
- Andrade, F.F.; Gomes, R.; Martins-Oliveira, I.; Dias, A.; Rodrigues, A.G.; Pina-Vaz, C. A Rapid Flow Cytometric Antimicrobial Susceptibility Assay (FASTvet) for Veterinary Use-Preliminary Data. Front. Microbiol. 2020, 11, 1944. [Google Scholar] [CrossRef]
- Jamerlan, A.; An, S.S.A.; Hulme, J. Advances in amyloid beta oligomer detection applications in Alzheimer’s disease. TrAC Trends Anal. Chem. 2020, 129, 115919. [Google Scholar] [CrossRef]
- Giau, V.V.; An, S.S.A.; Hulme, J. Recent advances in the treatment of pathogenic infections using antibiotics and nano-drug delivery vehicles. Drug Des. Devel. Ther. 2019, 13, 327–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paniel, N.; Noguer, T. Detection of Salmonella in food matrices, from conventional methods to recent aptamer-sensing technologies. Foods 2019, 8, 371. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Singh, A.K.; Senapati, D.; Fan, Z.; Ray, P.C. Bio-conjugated popcorn shaped gold nanoparticles for targeted photothermal killing of multiple drug resistant Salmonella DT104. J. Mater. Chem. 2011, 21, 17705–17709. [Google Scholar] [CrossRef]
- Jia, M.; Liu, Z.; Wu, C.; Zhang, Z.; Ma, L.; Lu, X.; Mao, Y.; Zhang, H. Detection of Escherichia coli O157:H7 and Salmonella enterica serotype Typhimurium based on cell elongation induced by beta-lactam antibiotics. Analyst 2019, 144, 4505–4512. [Google Scholar] [CrossRef]
- Wang, D.; He, P.; Wang, Z.; Li, G.; Majed, N.; Gu, A.Z. Advances in single cell Raman spectroscopy technologies for biological and environmental applications. Curr. Opin. Biotechnol. 2020, 64, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Hamme, A.T. Targeted highly sensitive detection/eradication of multidrug resistant Salmonella DT104 through gold nanoparticle-SWCNT bioconjugated nanohybrids. Analyst 2014, 139, 3702–3705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, D.; Wieland, K.; Qiu, L.; Neumann-Cip, A.C.; Magistro, G.; Stief, C.; Wieser, A.; Haisch, C. Heteroresistant Bacteria Detected by an Extended Raman-Based Antibiotic Susceptibility Test. Anal. Chem. 2020, 92, 8722–8731. [Google Scholar] [CrossRef]
- Cui, L.; Chen, P.; Chen, S.; Yuan, Z.; Yu, C.; Ren, B.; Zhang, K. In Situ Study of the Antibacterial Activity and Mechanism of Action of Silver Nanoparticles by Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2013, 85, 5436–5443. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, H.Z.; Zhu, X.; Su, J.Q.; Ren, B.; Zhu, Y.G.; Cui, L. Rapid Antibiotic Susceptibility Testing of Pathogenic Bacteria Using Heavy-Water-Labeled Single-Cell Raman Spectroscopy in Clinical Samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef]
- Cui, L.; Yang, K.; Li, H.Z.; Zhang, H.; Su, J.Q.; Paraskevaidi, M.; Martin, F.L.; Ren, B.; Zhu, Y.G. Functional Single-Cell Approach to Probing Nitrogen-Fixing Bacteria in Soil Communities by Resonance Raman Spectroscopy with 15N2 Labeling. Anal. Chem. 2018, 90, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhang, D.D.; Yang, K.; Zhang, X.; Zhu, Y.G. Perspective on Surface-Enhanced Raman Spectroscopic Investigation of Microbial World. Anal. Chem. 2019, 91, 15345–15354. [Google Scholar] [CrossRef]
- Bautista, D.A. ATP Bioluminescence: Application in Meat Industry. Encycl. Food Microbiol. 2014, 97–104. [Google Scholar] [CrossRef]
- Mempin, R.; Tran, H.; Chen, C.; Gong, H.; Ho, K.K.; Lu, S. Release of extracellular ATP by bacteria during growth. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihssen, J.; Jovanoic, N.; Sirec, T.; Spitz, U. Real-time monitoring of extracellular ATP in bacterial cultures using thermostable luciferase. PLoS ONE 2021, 16, e0244200. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L. New Rapid Bioassay of Gentamicin Based on Luciferase Assay of Extracellular ATP in Bacterial Cultures. Antimicrob. Agents Chemother. 1978, 14, 812–816. [Google Scholar] [CrossRef] [Green Version]
- Duan, N.; Sun, W.; Wu, S.; Liu, L.; Hun, X.; Wang, Z. Aptamer-based F0F1-ATPase biosensor for Salmonella typhimurium detection. Sens. Actuators B Chem. 2018, 255, 2582–2588. [Google Scholar] [CrossRef]
- Yin, M.; Wu, C.; Li, H.; Jia, Z.; Deng, Q.; Wang, S.; Zhang, Y. Simultaneous Sensing of Seven Pathogenic Bacteria by Guanidine-Functionalized Upconversion Fluorescent Nanoparticles. ACS Omega 2019, 4, 8953–8959. [Google Scholar] [CrossRef] [Green Version]
- Zeinhom, M.M.A.; Wang, Y.; Sheng, L.; Du, D.; Li, L.; Zhu, M.J.; Lin, Y. Smart phone based immunosensor coupled with nanoflower signal amplification for rapid detection of Salmonella Enteritidis in milk, cheese and water. Sens. Actuators B Chem. 2018, 261, 75–82. [Google Scholar] [CrossRef]
- Pramanik, A.; Davis, D.; Patibandla, S.; Begum, S.; Ray, P.; Gates, K.; Gao, Y.; Chandra Ray, P. A WS2-gold nanoparticle heterostructure-based novel SERS platform for the rapid identification of antibiotic-resistant pathogens. Nanoscale Adv. 2020, 2025–2033. [Google Scholar] [CrossRef] [Green Version]
- Zeni, L.; Perri, C.; Cennamo, N.; Arcadio, F.; D’Agostino, G.; Salmona, M.; Beeg, M.; Gobbi, M. A portable optical-fibre-based surface plasmon resonance biosensor for the detection of therapeutic antibodies in human serum. Sci. Rep. 2020, 10, 11154. [Google Scholar] [CrossRef]
- Xu, Y.; Luo, Z.; Chen, J.; Huang, Z.; Wang, X.; An, H.; Duan, Y. ω-Shaped Fiber-Optic Probe-Based Localized Surface Plasmon Resonance Biosensor for Real-Time Detection of Salmonella Typhimurium. Anal. Chem. 2018, 90, 13640–13646. [Google Scholar] [CrossRef] [PubMed]
- Paracini, N.; Clifton, L.A.; Skoda, M.W.A.; Lakey, J.H. Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility. Proc. Natl. Acad. Sci. USA 2018, 115, E7587–E7594. [Google Scholar] [CrossRef] [Green Version]
- Zafiu, C.; Hussain, Z.; Kupcu, S.; Masutani, A.; Kilickiran, P.; Skinner, E.K. Liquid crystals as optical amplifiers for bacterial detection. Biosens. Bioelectron. 2016, 80, 161–170. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredborg, M.; Andersen, K.R.; Jørgensen, E.; Droce, A.; Olesen, T.; Jensen, B.B.; Rosenvinge, F.S.; Sondergaard, T.E. Real-time optical antimicrobial susceptibility testing. J. Clin. Microbiol. 2013, 51, 2047–2053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Jing, W.; Iriya, R.; Yang, Y.; Syal, K.; Mo, M.; Grys, T.E.; Haydel, S.E.; Wang, S.; Tao, N. Phenotypic Antimicrobial Susceptibility Testing with Deep Learning Video Microscopy. Anal. Chem. 2018, 90, 6314–6322. [Google Scholar] [CrossRef]
- Bolostky, A.; Muralidhan, R.; Butler, D.; Root, K.; Murray, W.; Liu, Z.; Ebrahimi, A. Organic redox-active crystallinelayers for reagent-free electrochemical antibiotic susceptibility testing (ORACLE-AST). Biosens. Bioelectron. 2021, 172, 112615. [Google Scholar]
- Gao, Y.; Ryu, J.; Liu, L.; Choi, S. A simple, inexpensive, and rapid method to assess antibiotic effectiveness against exoelectrogenic bacteria. Biosens. Bioelectron. 2020, 168, 112518. [Google Scholar] [CrossRef]
- Rao, R.P.; Sharma, S.; Mehrotra, T.; Das, R.; Kumar, R.; Singh, R.; Roy, I.; Basu, T. Rapid Electrochemical Monitoring of Bacterial Respiration for Gram-Positive and Gram-Negative Microbes: Potential Application in Antimicrobial Susceptibility Testing. Anal. Chem. 2020, 92, 4266–4274. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Narang, J.; Pundir, C.S.; Pilloton, R.; Khanuja, M. Morphology-Preferable MoSe2 Nanobrooms as a Sensing Platform for Highly Selective Apta-Capturing of Salmonella Bacteria. ACS Omega 2018, 3, 13020–13027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besant, J.D.; Sargent, E.H.; Kelley, S.O. Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip 2015, 15, 2799–2807. [Google Scholar] [CrossRef]
- Ren, Y.; Ji, J.; Sun, J.; Pi, F.; Zhang, Y.; Sun, X. Rapid detection of antibiotic resistance in Salmonella with screen printed carbon electrodes. J. Solid State Electrochem. 2020, 24, 1539–1549. [Google Scholar] [CrossRef]
- Bolostky, A.; Muralidhan, R.; Butler, D.; Root, K.; Murray, W.; Liu, Z.; Ebrahimi, A. Reagent-Free Electrochemical Monitoring of Bacterial Viability Using Novel Organic Crystals Deposited on Flexible Substrates. Meet. Abstr. MA 2020, 2, 2840. [Google Scholar]
- Liu, X.; Zheng, S.; Hu, Y.; Li, Z.; Luo, F.; He, Z. Electrochemical Immunosensor Based on the Chitosan-Magnetic Nanoparticles for Detection of Tetracycline. Food Anal. Methods 2016, 9, 2972–2978. [Google Scholar] [CrossRef]
- Stevenson, H.S.; Shetty, S.S.; Thomas, N.J.; Dhamu, V.N.; Bhide, A.; Prasad, S. Ultrasensitive and Rapid-Response Sensor for the Electrochemical Detection of Antibiotic Residues within Meat Samples. ACS Omega 2019, 4, 6324–6330. [Google Scholar] [CrossRef]
- Nair, D.V.T.; Venkitanarayanan, K.; Johny, A.K. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 2018, 7, 167. [Google Scholar] [CrossRef] [Green Version]
- Phothaworn, P.; Supokaivanich, R.; Lim, J.; Klumpp, J.; Imam, M.; Kutter, E.; Galyov, E.E.; Dunne, M.; Korbsrisate, S. Development of a broad-spectrum Salmonella phage cocktail containing Viunalike and Jerseylike viruses isolated from Thailand. Food Microbiol. 2020, 92, 103586. [Google Scholar] [CrossRef] [PubMed]





| Salmonella Serotypes | Sensing Method | Sample Matrix | Analysis Time (min) | Detection Limit (CFU/mL) | Reference |
|---|---|---|---|---|---|
| S. Typhimurium DT104 | SERS | Assay media | 30 | 105 | [71] |
| S. Typhimurium DT104 | SERS | Assay media | 15 | 105 | [74] |
| S. Typhimurium | SERS | H2O & milk | 120 | 20 | [72] |
| S. enterica | Raman Spectroscopy | Urine | 150 | n/a | [77] |
| S. Enteritidis, | Fluorescence | Water, milk, and beef | 30 | 2.0 × 102 | [86] |
| S. Typhimurium DT104 and S. Typhi | SERS | Assay media | 120 | 100 | [87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Hulme, J.P. Recent Advances in the Detection of Antibiotic and Multi-Drug Resistant Salmonella: An Update. Int. J. Mol. Sci. 2021, 22, 3499. https://doi.org/10.3390/ijms22073499
Wu S, Hulme JP. Recent Advances in the Detection of Antibiotic and Multi-Drug Resistant Salmonella: An Update. International Journal of Molecular Sciences. 2021; 22(7):3499. https://doi.org/10.3390/ijms22073499
Chicago/Turabian StyleWu, Siying, and John P. Hulme. 2021. "Recent Advances in the Detection of Antibiotic and Multi-Drug Resistant Salmonella: An Update" International Journal of Molecular Sciences 22, no. 7: 3499. https://doi.org/10.3390/ijms22073499
APA StyleWu, S., & Hulme, J. P. (2021). Recent Advances in the Detection of Antibiotic and Multi-Drug Resistant Salmonella: An Update. International Journal of Molecular Sciences, 22(7), 3499. https://doi.org/10.3390/ijms22073499