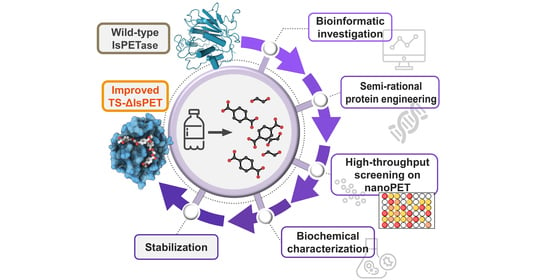
An Efficient Protein Evolution Workflow for the Improvement of Bacterial PET Hydrolyzing Enzymes
Abstract
:1. Introduction
2. Results
2.1. In Silico Analysis of the Interaction between ΔIsPET and PET
2.2. Production of Evolved ΔIsPET Variants
2.3. Biochemical Properties of Single-Point ΔIsPET Variants
2.4. Enhancement of the Thermal Stability of the F238A-ΔIsPET Variant
2.5. Kinetic Parameters of ΔIsPET Variants on PET Nanoparticles
2.6. MD Analysis of ΔIsPET Variants
2.7. Biodegradation of PET Microplastics by TS-ΔIsPET
3. Discussion
4. Materials and Methods
4.1. In Silico Analyses
4.2. Preparation of PET Nanoparticles
4.3. Cloning, Expression, and Purification of ΔIsPET
4.4. Site-Saturation Mutagenesis and Generation of Mutant Libraries
4.5. High-Throughput Screening for Evolved ΔIsPET Variants
4.6. Activity Assays
4.7. Thermal Stability of ΔIsPET Variants
4.8. Enzymatic Bioconversion of PET Microparticles
4.9. Determination of the Adsorption of ΔIsPET Variants to PET Microparticles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rochman, C.M.; Hoh, E.; Hentschel, B.T.; Kaye, S. Classify plastic waste as hazardous (types of externalities caused by consumption of plastic bags). Environ. Sci. Technol. 2013, 47, 1646–1654. [Google Scholar] [PubMed]
- Porta, R. Anthropocene, the plastic age and future perspectives. FEBS Open Bio 2021, 11, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Oeser, T.; Barth, M.; Weigl, N.; Lübs, A.; Schulz-Siegmund, M.; Hacker, M.C.; Zimmermann, W. Turbidimetric analysis of the enzymatic hydrolysis of polyethylene terephthalate nanoparticles. J. Mol. Catal. B Enzym. 2014, 103, 72–78. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current status and application aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Pirillo, V.; Pollegioni, L.; Molla, G. Analytical methods for the investigation of enzyme-catalyzed degradation of polyethylene terephthalate. FEBS J. 2021, 288, 4730–4745. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Clarke, D.J.; Dobson, A.D.W. Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Front. Microbiol. 2020, 11, 1–23. [Google Scholar] [CrossRef]
- Leitão, A.L.; Enguita, F.J. Structural insights into carboxylic polyester-degrading enzymes and their functional depolymerizing neighbors. Int. J. Mol. Sci. 2021, 22, 2332. [Google Scholar] [CrossRef] [PubMed]
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.Y.; Lee, S.H.; Hong, H.; Kim, K.J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzyme Microb. Technol. 2020, 141, 109656. [Google Scholar] [CrossRef]
- Meng, X.; Yang, L.; Liu, H.; Li, Q.; Xu, G.; Zhang, Y.; Guan, F.; Zhang, Y.; Zhang, W.; Wu, N.; et al. Protein engineering of stable IsPETase for PET plastic degradation by Premuse. Int. J. Biol. Macromol. 2021, 180, 667–676. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takanaha, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethyleneterephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X.; et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.W.; Ma, J.; Zheng, Y.; Ko, T.P.; Xu, L.; Cheng, Y.S.; Chen, C.C.; Guo, R.T. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Han, X.; Ko, T.P.; Liu, W.; Guo, R.T. Structural studies reveal the molecular mechanism of PETase. FEBS J. 2018, 285, 3717–3723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boneta, S.; Arafet, K.; Moliner, V. QM/MM study of the enzymatic biodegradation mechanism of polyethylene terephthalate. J. Chem. Inf. Model. 2021, 61, 3041–3051. [Google Scholar] [CrossRef] [PubMed]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kan, Y.; He, L.; Luo, Y.; Bao, R. IsPETase is a novel biocatalyst for poly(ethylene terephthalate) (PET) hydrolysis. ChemBioChem 2021, 22, 1706–1716. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef] [Green Version]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- da Costa, C.H.S.; dos Santos, A.M.; Alves, C.N.; Martí, S.; Moliner, V.; Santana, K.; Lameira, J. Assessment of the PETase conformational changes induced by poly(ethylene terephthalate) binding. Proteins Struct. Funct. Bioinforma. 2021, 89, 1340–1352. [Google Scholar] [CrossRef]
- Ma, Y.; Yao, M.; Li, B.; Ding, M.; He, B.; Chen, S.; Zhou, X.; Yuan, Y. Enhanced poly(ethylene terephthalate) hydrolase activity by protein engineering. Engineering 2018, 4, 888–893. [Google Scholar] [CrossRef]
- Pfaff, L.; Breite, D.; Badenhorst, C.P.S.; Bornscheuer, U.T.; Wei, R. Fluorimetric high-throughput screening method for polyester hydrolase activity using polyethylene terephthalate nanoparticles. Methods Enzymol. 2021, 648, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Scandola, M.; Focarete, M.L.; Frisoni, G. Simple kinetic model for the heterogeneous enzymatic hydrolysis of natural poly(3-hydroxybutyrate). Macromolecules 1998, 31, 3846–3851. [Google Scholar] [CrossRef]
- Fecker, T.; Galaz-Davison, P.; Engelberger, F.; Narui, Y.; Sotomayor, M.; Parra, L.P.; Ramírez-Sarmiento, C.A. Active site flexibility as a hallmark for efficient PET degradation by I. sakaiensis PETase. Biophys. J. 2018, 114, 1302–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, J.P. Protein flexibility and stiffness enable efficient enzymatic catalysis. J. Am. Chem. Soc. 2019, 141, 3320–3331. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Song, C.; Gräsing, D.; Schneider, T.; Bielytskyi, P.; Böttcher, D.; Matysik, J.; Bornscheuer, U.T.; Zimmermann, W. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 2019, 10, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Falkenstein, P.; Wei, R.; Matysik, J.; Song, C. Mechanistic investigation of enzymatic degradation of polyethylene terephthalate by nuclear magnetic resonance. Methods Enzymol. 2021, 648, 231–252. [Google Scholar] [CrossRef]
- Badino, S.F.; Bååth, J.A.; Borch, K.; Jensen, K.; Westh, P. Adsorption of enzymes with hydrolytic activity on polyethylene terephthalate. Enzyme Microb. Technol. 2021, 152, 109937. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, 522–525. [Google Scholar] [CrossRef]
- Zhang, N.; Zhao, H. Enriching screening libraries with bioactive fragment space. Bioorg. Med. Chem. Lett. 2016, 26, 3594–3597. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: The prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Longhi, S.; Cambillau, C. Structure-activity of cutinase, a small lipolytic enzyme. Biochim. Biophys. Acta 1999, 1441, 185–196. [Google Scholar] [CrossRef]
- Oh, C.; Doohun Kim, T.; Kim, K.K. Carboxylic ester hydrolases in bacteria: Active site, structure, function and application. Crystals 2019, 9, 597. [Google Scholar] [CrossRef] [Green Version]
- Sadiq, S.K.; Coveney, P.V. Computing the role of near attack conformations in an enzyme-catalyzed nucleophilic bimolecular reaction. J. Chem. Theory Comput. 2015, 11, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Miller, B.R.; Mcgee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J Chem Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, M.; Moreno Castillo, E.; Ihling, C.H.; Iacobucci, C.; Wilde, V.; Hellmuth, A.; Hoehenwarter, W.; Samodelov, S.L.; Zurbriggen, M.D.; Kastritis, P.L.; et al. Flexibility of intrinsically disordered degrons in AUX/IAA proteins reinforces auxin co-receptor assemblies. Nat. Commun. 2020, 11, 2277. [Google Scholar] [CrossRef]
- Ubbiali, D.; Orlando, M.; Kovačič, M.; Iacobucci, C.; Semrau, M.S.; Bajc, G.; Fortuna, S.; Ilc, G.; Medagli, B.; Oloketuyi, S.; et al. An anti-HER2 nanobody binds to its antigen HER2 via two independent paratopes. Int. J. Biol. Macromol. 2021, 182, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pupko, T.; Bell, R.E.; Mayrose, I.; Glaser, F.; Ben-Tal, N. Rate4Site: An algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 2002, 18, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Mörtl, M.; Diederichs, K.; Welte, W.; Molla, G.; Motteran, L.; Andriolo, G.; Pilone, M.S.; Pollegioni, L. Structure-function correlation in glycine oxidase from Bacillus subtilis. J. Biol. Chem. 2004, 279, 29718–29727. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Sehata, S.; Suzuki, R.; Koumoto, K. Increased yield of Β-glucosidase-catalyzed hydrolysis reactions in the presence of betaine-type metabolite analog. Bioprocess Biosyst. Eng. 2017, 40, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Herzog, K.; Müller, R.J.; Deckwer, W.D. Mechanism and kinetics of the enzymatic hydrolysis of polyester nanoparticles by lipases. Polym. Degrad. Stab. 2006, 91, 2486–2498. [Google Scholar] [CrossRef]
- Caldinelli, L.; Molla, G.; Bracci, L.; Lelli, B.; Pileri, S.; Cappelletti, P.; Sacchi, S.; Pollegioni, L. Effect of ligand binding on human D-amino acid oxidase: Implications for the development of new drugs for schizophrenia treatment. Protein Sci. 2010, 19, 1500–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Wang, D.; Wei, N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022, 40, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, J.K.; Cha, H.G.; Kang, M.J.; Lee, H.S.; Khang, T.U.; Yun, E.J.; Lee, D.H.; Song, B.K.; Park, S.J.; et al. Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406. [Google Scholar] [CrossRef]






| Variants | kτ (min−1) | KA (mL mg−1) | R2 |
|---|---|---|---|
| ΔIsPET | 0.076 ± 0.004 | 37.95 ± 5.20 | 0.99 |
| F238A-ΔIsPET | 0.124 ± 0.003 | 75.38 ± 12.14 | 0.99 |
| TS-ΔIsPET | 0.098 ± 0.002 | 95.51 ± 11.90 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pirillo, V.; Orlando, M.; Tessaro, D.; Pollegioni, L.; Molla, G. An Efficient Protein Evolution Workflow for the Improvement of Bacterial PET Hydrolyzing Enzymes. Int. J. Mol. Sci. 2022, 23, 264. https://doi.org/10.3390/ijms23010264
Pirillo V, Orlando M, Tessaro D, Pollegioni L, Molla G. An Efficient Protein Evolution Workflow for the Improvement of Bacterial PET Hydrolyzing Enzymes. International Journal of Molecular Sciences. 2022; 23(1):264. https://doi.org/10.3390/ijms23010264
Chicago/Turabian StylePirillo, Valentina, Marco Orlando, Davide Tessaro, Loredano Pollegioni, and Gianluca Molla. 2022. "An Efficient Protein Evolution Workflow for the Improvement of Bacterial PET Hydrolyzing Enzymes" International Journal of Molecular Sciences 23, no. 1: 264. https://doi.org/10.3390/ijms23010264
APA StylePirillo, V., Orlando, M., Tessaro, D., Pollegioni, L., & Molla, G. (2022). An Efficient Protein Evolution Workflow for the Improvement of Bacterial PET Hydrolyzing Enzymes. International Journal of Molecular Sciences, 23(1), 264. https://doi.org/10.3390/ijms23010264