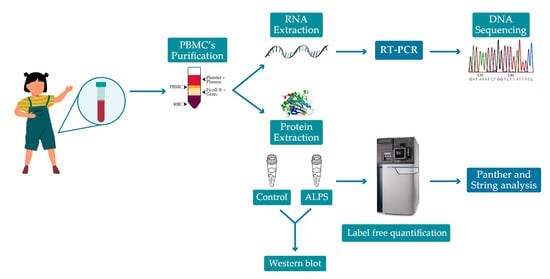
Differential Expression of Proteins in an Atypical Presentation of Autoimmune Lymphoproliferative Syndrome
Abstract
:1. Introduction
2. Results
2.1. Clinical Features of the Patients
2.2. Molecular Analysis of Fas, FasL and Casp10 Genes
2.3. Proteomic Analysis of PBMCs
2.4. Identification of Biological Pathways of Detected Proteins
2.5. Protein-Protein Interactions
2.6. E2F7 Study
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. RNA Analysis
4.3. Label-Free Mass Spectrometry
4.3.1. Sample Preparation
4.3.2. Quantitative Mass Spectrometry and Data Analysis
4.3.3. Bioinformatics Analysis
4.3.4. Western Blotting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Price, S.; Shaw, P.A.; Seitz, A.; Joshi, G.; Davis, J.; Niemela, J.E.; Perkins, K.; Hornung, R.L.; Folio, L.; Rosenberg, P.S.; et al. Natural history of autoimmune lymphoproliferative syndrome associated with FAS gene mutations. Blood 2014, 123, 1989–1999. [Google Scholar] [CrossRef] [PubMed]
- Straus, S.E.; Jaffe, E.S.; Puck, J.M.; Dale, J.K.; Elkon, K.B.; Rösen-Wolff, A.; Peters, A.M.J.; Sneller, M.C.; Hallahan, C.W.; Wang, J.; et al. The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood 2001, 98, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Consonni, F.; Gambineri, E.; Favre, C. ALPS, FAS, and beyond: From in born errors of immunity to acquired immunodeficiencies. Annal. Hematol. 2022, 101, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Rieux-Laucat, F.; Magérus-Chatinet, A.; Neven, B. The autoimmune lymphoproliferative syndrome with defective FAS or FAS-ligand functions. J. Clin. Immunol. 2018, 38, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Spergel, A.R.; Walkovich, K.; Price, S.; Niemela, J.E.; Wright, D.; Fleisher, T.; Rao, V.K. Autoimmune lymphoproliferative syndrome misdiagnosed as hemophagocytic lymphohistiocytosis. Pediactrics 2013, 132, e1440–e1444. [Google Scholar] [CrossRef] [Green Version]
- Colino, C.G. Avances en el conocimiento y manejo del síndrome linfoproliferativo autoinmune. An. Pediatr. 2014, 80, 122.e1–122.e7. [Google Scholar] [CrossRef]
- Miano, M.; Cappelli, E.; Pezzulla, A.; Venè, R.; Grossi, A.; Terranova, P.; Palmisani, E.; Maggiore, R.; Guardo, D.; Lanza, T.; et al. FAS-mediated apoptosis impairment in patients with ALPS/ALPS-like phenotype carrying variants on CASP10 gene. Br. J. Haematol. 2019, 187, 502–508. [Google Scholar] [CrossRef]
- Oliveira, J.B.; Bleesing, J.J.; Dianzani, U.; Fleisher, T.A.; Jaffe, E.S.; Lenardo, M.J.; Rieux-Laucat, F.; Siegel, R.M.; Su, H.C.; Su, H.C.; et al. Revised diagnostic criteria and classification for the autoimmune lymphoproliferative syndrome (ALPS): Report from the 2009 NIH International Workshop. Blood 2010, 116, e35–e40. [Google Scholar] [CrossRef]
- Teachey, D.T. New advances in the diagnosis and treatment of autoimmune lymphoproliferative syndrome. Curr. Opin. Pediatr. 2012, 24, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ashoka, A.; Bilal, J. Generalized lymphoadenopathy with cytopenias. Am. J. Med. 2021, 134, e490–e491. [Google Scholar] [CrossRef]
- Lee, C.L.; Jiang, P.P.; Sit, W.H.; Wan, J.M.F. Proteome of human T lymphocytes with treatment of cyclosporine and polysaccharopeptide: Analysis of significant proteins that manipulate T cells proliferation and immunosuppression. Int. Immunopharmacol. 2007, 7, 1311–1324. [Google Scholar] [CrossRef] [PubMed]
- Gulli, F.; Basile, U.; Gragnani, L.; Fognani, E.; Napodano, C.; Colacicco, L.; Miele, L.; de Matthaeis, N.; Cattani, P.; Zignego, A.L.; et al. Autoimmunity and lymphoproliferation markers in naïve HCV-RNA positive patients without clinical evidences of autoimmune/lymphoproliferative disorders. Dig. Liver Dis. 2016, 48, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Sobh, A.; Crestani, E.; Cangemi, B.; Kane, J.; Chou, J.; Pai, A.Y.; Notarangelo, L.D.; Al-Herz, W.; Geha, R.S.; Massaad, M.J. Autoimmune lymphoproliferative syndrome caused by a homozygous FasL mutation that disrupts FasL assembly. J. Allergy Clin. Immunol. 2016, 137, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Hsu, A.P.; Vacek, M.M.; Zheng, L.; Schäffer, A.A.; Dale, J.K.; Davis, J.; Fischer, R.E.; Straus, S.E.; Boruchov, D.; et al. Genetic alterations in caspase-10 may be causative or protective in autoimmune lymphoproliferative syndrome. Hum. Genet. 2006, 119, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Drappa, J.; Vaishnaw, A.K.; Sullivan, K.E.; Chu, J.L.; Elkon, K.B. Fas gene mutations in the Canale-Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N. Engl. J. Med. 1996, 335, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, P.; Sun, X.; Luo, Y.; Wang, X.; Ma, D.; Wu, J. Cloning and characterization of a novel caspase-10 isoform that activates NF-κB activity. Biochim. Biophys. Acta 2007, 1770, 1528–1537. [Google Scholar] [CrossRef]
- Wang, J.; Lobito, A.; Chan, F.K.-M.; Dale, J.; Sneller, M.; Yao, X.; Puck, J.M.; Straus, S.E.; Lenardo, M.J. Inherited human caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell 1999, 98, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, S.I.; Mazza, C.; Moratto, D.; Ramenghi, U.; Caorsi, R.; Gattorno, M.; Badolato, R. Atypical presentation of autoimmune lymphoproliferative syndrome due to CASP10 mutation. Immunol. Lett. 2016, 177, 22–24. [Google Scholar] [CrossRef]
- Nabhani, S.; Hönscheid, A.; Oommen, P.T.; Fleckenstein, B.; Schaper, J.; Kuhlen, M.; Laws, H.; Borkhardt, A.; Fischer, U. A novel homozygous Fas ligand mutation leads to early protein truncation, abrogation of death receptor and reverse signaling and a severe form of the autoimmune lymphoproliferative syndrome. Clin. Immunol. 2014, 155, 231–237. [Google Scholar] [CrossRef]
- Bi, L.L.; Pan, G.; Prescott, A.T.; Zheng, L.; Dale, J.K.; Makris, C.; Reddy, V.; McDonald, J.M.; Siegel, R.M.; Puck, J.M.; et al. Dominant inhibition of Fas ligand-mediated apoptosis due to a heterozygous mutation associated with autoimmune lymphoproliferative syndrome (ALPS) Type Ib. BMC Med. Genet. 2007, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Del-Rey, M.; Ruiz-Contreras, J.; Bosque, A.; Calleja, S.; Gomez-Rial, J.; Roldan, E.; Morales, P.; Serrano, A.; Anel, A.; Paz-Artal, E.; et al. A homozygous Fas ligand gene mutation in a patient causes a new type of autoimmune lymphoproliferative syndrome. Blood 2006, 108, 1306–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhu, Z.; Bai, C.; Sun, H.; Wang, X. Proteomic profiling of lymphocytes in autoimmunity, inflammation and cancer. J. Transl. Med. 2014, 12, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, G.H.M.F.; Guest, P.C.; Martins-De-Souza, D. LC-MSE, Multiplex MS/MS, Ion mobility, and Label-Free Quantitation in Clinical Proteomics. Methods Mol. Biol. 2017, 1546, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ran, C.; Li, E.; Gordon, F.; Comstock, G.; Siddiqui, H.; Cleghorn, W.; Chen, H.; Kornacker, K.; Liu, C.; et al. Synergistic Function of E2F7 and E2F8 Is Essential for Cell Survival and Embryonic Development. Dev. Cell 2008, 14, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Bandura, J.L.; Jiang, H.; Nickerson, D.W.; Edgar, B.A. The Molecular Chaperone Hsp90 Is Required for Cell Cycle Exit in Drosophila melanogaster. PLoS Genet. 2013, 9, e1003835. [Google Scholar] [CrossRef] [Green Version]
- Westendorp, B.; Mokry, M.; Koerkamp, M.J.A.G.; Holstege, F.C.P.; Cuppen, E.; Bruin, A.D. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 2012, 40, 3511–3523. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Grossmann, P.; Padi, M.; DeCaprio, J.A. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016, 44, 6070–6086. [Google Scholar] [CrossRef]
- Lu, R.; Markowetz, F.; Unwin, R.D.; Leek, J.T.; Airoldi, E.M.; MacArthur, B.D.; Lachmann, A.; Rozov, R.; Ma’ayan, A.; Boyer, L.A.; et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 2009, 462, 358–364. [Google Scholar] [CrossRef] [Green Version]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian geme expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.G.; Cao, Y.-B.; Cao, Y.-Y.; Zhang, J.-D.; An, M.-M.; Wang, Y.; Gao, P.-H.; Yan, L.; Xu, Y.; Jiang, Y.-Y. Altered protein profile of lymphocytes in an antigen-specific model of colitis: A comparative proteomic study. Inflamm. Res. 2007, 56, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Haendel, M.; Vasilevsky, N.; Unni, D.; Bologa, C.; Harris, N.; Rhem, H.; Hamosh, A.; Baynam, G.; Groza, T.; McMurry, J.; et al. How many rare diseases are there? Nat. Rev. Drug Discov. 2020, 19, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braconi, D.; Bernardini, G.; Spiga, O.; Santucci, A. Leveraging proteomics in orphan disease research: Pitfalls and potential. Expert Rev. Proteom. 2021, 18, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Baldini, C.; Giusti, L.; Ciregia, F.; Valle, Y.D.; Giacomelli, C.; Donadio, E.; Ferro, F.; Galimberti, S.; Donati, V.; Bazzichi, L.; et al. Correspondence between salivary proteomic pattern and clinical course in primary Sjögren syndrome and non-Hodgkin’s lymphoma: A case report. J. Transl. Med. 2011, 9, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aqrawi, L.A.; Galtung, H.K.; Vestad, B.; Ovstebo, R.; Thiede, B.; Rusthen, S.; Young, A.; Guerreiro, E.M.; Utheim, T.P.; Chen, X.; et al. Identification of potential salivary and tear biomarkers in primary Sjögren’s syndrome, utilizing the extraction of extracellular vesicles and proteomics analysis. Arthritis Res. Ther. 2017, 19, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magagnotti, C.; Bachi, A.; Zerbini, G.; Fattore, E.; Fermo, I.; Riba, M.; Previtali, S.C.; Ferrari, M.; Andolfo, A.; Benedetti, S. Protein profiling reveals energy metabolism and cytoskeletal protein alterations in LMNA mutation carriers. Biochim. Biophys. Acta 2012, 1822, 970–979. [Google Scholar] [CrossRef] [Green Version]
- Dianzani, U.; Bragardo, M.; DiFranco, D.; Alliaudi, C.; Scagni, P.; Buonfiglio, D.; Redoglia, V.; Bonissoni, S.; Correra, A.; Dianzani, I.; et al. Deficiency of the Fas apoptosis pathway without Fas gene mutations in pediatric patients with autoimmunity/lymphoproliferation. Blood 1997, 89, 2871–2879. [Google Scholar] [CrossRef]
- Infante, A.J.; Britton, H.A.; DeNapoli, T.; Middelton, L.A.; Lenardo, M.J.; Jackson, C.E.; Wang, J.; Fleisher, T.; Straus, S.E.; Puck, J.M. The clinical spectrum in a large kindred with autoimmune lymphoproliferative syndrome caused by a Fas mutation that impairs lymphocyte apoptosis. J. Pediatr. 1998, 133, 629–633. [Google Scholar] [CrossRef]
- Heller, T.; Asif, A.R.; Petrova, D.T.; Doncheva, Y.; Wieland, E.; Oellerich, M.; Shipkova, M. Differential proteomic analysis of lymphocytes treated ith mycophenolic acid reveals caspase 3-induced cleavage of Rho GDP dissociation inhibior 2. Ther. Drug Monit. 2009, 31, 211–217. [Google Scholar] [CrossRef]
- Coutts, A.S.; Munro, S.; Thangue, N.B.L. Functional interplay between E2F7 and ribosomal rRNA gene transcription regulates protein synthesis. Cell Death Dis. 2018, 9, 577. [Google Scholar] [CrossRef] [Green Version]
- Zalmas, L.-P.; Zhao, X.; Graham, A.; Fisher, R.; Reilly, C.; Coutts, A.S.; Thangue, N.B.L. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008, 9, 252–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazar-Rethinam, M.; Cameron, S.R.; Dahler, A.L.; Endo-Munoz, L.B.; Smith, L.; Rickwood, D.; Saunders, N.A. Loss of E2F7 expression is an early event in squamous differentiation and causes derepression of the key differentiation activator Sp1. J. Invest. Dermatol. 2011, 131, 1077–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitxelena, J.; Apraiz, A.; Vallejo-Rodríguez, J.; García-Santisteban, I.; Fullaondo, A.; Alvarez-Fernández, M.; Malumbres, M.; Zubiaga, A.M. An E2F7-dependent transcriptional program modulates DNA damage repair and genomic stability. Nucleic Acids Res. 2018, 46, 4546–4559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer. 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Thurlings, I.; Martínez-López, L.M.; Westendorp, B.; Zijp, M.; Kuiper, R.; Tooten, P.; Kent, L.N.; Leone, G.; Vos, H.J.; Burgering, B.; et al. Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene 2017, 36, 829–839. [Google Scholar] [CrossRef] [Green Version]
- Zalmas, L.P.; Coutts, A.S.; Helleday, T.; Thangue, N.B.L. E2F-7 couples DNA damage-dependent transcription with the DNA repair process. Cell Cycle 2013, 12, 3037–3051. [Google Scholar] [CrossRef]
- Kleiveland, C.R. Peripheral blood mononuclear cells. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 161–167. [Google Scholar] [CrossRef]
- Ashley, D.M.; Riffkin, C.D.; Muscat, A.M.; Knight, M.J.; Kaye, A.H.; Novak, U.; Hawkins, C.J. Caspase 8 is absent or low in many ex vivo gliomas. Cancer 2005, 104, 1487–1496. [Google Scholar] [CrossRef]
- Park, W.S.; Lee, J.H.; Shin, M.S.; Park, J.Y.; Kim, H.S.; Lee, J.H.; Kim, Y.S.; Lee, S.N.; Xiao, W.; Park, C.H.; et al. Inactivating mutations of the caspase-10 gene in gastric cancer. Oncogene 2002, 21, 2919–2995. [Google Scholar] [CrossRef] [Green Version]
- Clements, K.E.; Thakar, T.; Nicolae, C.M.; Liang, X.; Wang, H.G.; Moldovan, G.L. Loss of E2F7 confers resistance to poly-ADP-ribose polymerase (PARP) inhibitors in BRCA2-deficient cells. Nucleic Acids Res. 2018, 46, 8898–8907. [Google Scholar] [CrossRef]
- Yang, F.; Xu, Y.P.; Li, J.; Duan, S.S.; Fu, Y.J.; Zhang, Y.; Zhao, Y.; Qiao, W.; Chen, Q.; Geng, Y.; et al. Cloning and characterization of a novel intracellular protein p48.2 that negatively regulates cell cycle progression. Int. J. Biochem. Cell Biol. 2009, 41, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Thadikkaran, L.; Rufer, N.; Benay, C.; Crettaz, D.; Tissot, J.D. Methods for Human CD8+ T Lymphocyte Proteome Analysis. In Functional Proteomics; Humana Press: Totowa, NJ, USA, 2008; p. 484. [Google Scholar] [CrossRef]
- Zuccoli, G.S.; Martins-de-Souza, D.; Guest, P.C.; Rehen, S.K.; Nascimento, J.M. Combining patient-reprogrammed neural cells and proteomics as a model to study psychiatric disorders. Adv. Exp. Med. Biol. 2017, 974, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Castro, E.; Souza, G.H.M.F.; Delgadillo-Álvarez, D.M.; Ramírez-Reyes, L.; Torres-Huerta, A.; Velasco-Suarez, A.; Cruz-Cruz, C.; Hernández-Hernández, J.M.; Tapia-Ramírez, J. Quantitative Proteomic Analysis of MARC-145 Cells Infected with a Mexican Porcine Reproductive and Respiratory Syndrome Virus Strain Using a Label-Free Based DIA approach. J. Am. Soc. Mass Spectrom. 2020, 31, 1302–1312. [Google Scholar] [CrossRef]
- Li, G.Z.; Vissers, J.P.C.; Silva, J.C.; Golick, D.; Gorenstein, M.V.; Geromanos, S.J. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 2009, 9, 1696–1719. [Google Scholar] [CrossRef] [PubMed]
- Geromanos, S.J.; Hughes, C.; Golick, D.; Ciavarini, S.; Gorestein, M.V.; Richardson, K.; Hoyes, J.B.; Vissers, J.P.C.; Langridge, J.I. Simulating and validating proteomics data and search results. Proteomics 2011, 11, 1189–1211. [Google Scholar] [CrossRef] [PubMed]
- Valentine, S.J.; Ewing, M.A.; Dilger, J.M.; Glover, M.S.; Geromanos, S.J.; Hughes, C.; Clemmer, D.E. Using ion mobility data to improve peptide identification: Intrinsic Amino Acid Size Parameters. J. Proteome Res. 2011, 10, 2318–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, 442–450. [Google Scholar] [CrossRef]








| Patient 1 | Patient 2 | Normal Range | |
|---|---|---|---|
| Age (years) | 13 | 6 | |
| Age at diagnosis (years) | 10 | 6 | |
| Sex | Female | Male | |
| Clinical data | |||
| Splenomegaly/Hepatomegaly | + | + | |
| Lymphadenopathy | + | + | |
| Autoimmunity | |||
| Anemia | + | + | |
| Thrombopenia | + | + | |
| Neutropenia | + | + | |
| Laboratory results | |||
| Lymphocyte count (cells/µL) | 1450 | 3025 | 1800–2200 |
| T-cell (%) | |||
| CD3 | 88.5 | 77.9 | 60–85 |
| CD4 | 81.2 | 43.2 | 29–59 |
| CD8 | 5.28 | 28.9 | 19–48 |
| DN | 1.8 | 3.2 | <1% |
| B-cells (%) | |||
| CD19 | - | 12.7 | 8–20 |
| CD27+ | - | - | 8–45 |
| IgD-CD27+ (switching) | - | - | 5–25 |
| CD21Low | - | - | 1.5–9 |
| Serum immunoglobulins | |||
| (mg/dl) | |||
| IgG | 2557 | 3596 | 400–1000 |
| IgA | 452 | 200 | 68–385 |
| IgM | 292 | 328 | 60–264 |
| Plasma biomarkers (pg/mL) | |||
| IL-10 | - | - | |
| sFalL | - | - | |
| sCD25 (U/mL) | - | - | |
| Vitamin B12 (pg/mL) | - | 1412 | 180–914 |
| Treatment | Corticosteroids, spironolactone, ursodeoxycholic acid, azathioprine, Mycophenolate, Red Blood Cells transfusions | Corticosteroids, Mycophenolate |
| Fas | MLGIWTLLPLVLTSVARLSSKSVNAQVTDINSKGLELRKTVTTVETQNLEGLHHDGQFCH | 60 |
| Patient_1 | MLGIWTLLPLVLTSVARLSSKSVNAQVTDINSKGLELRKTVTTVETQNLEGLHHDGQFCH | 60 |
| Patient_2 | MLGIWTLLPLVLTSVARLSSKSVNAQVTDINSKGLELRKTVTTVETQNLEGLHHDGQFCH | 60 |
| Fas | KPCPPGERKARDCTVNGDEPDCVPCQEGKEYTDKAHFSSKCRRCRLCDEGHGLEVEINCT | 120 |
| Patient_1 | KPCPPGERKARDCTVNGDEPDCVPCQEGKEYTDKAHFSSKCRRCRLCDEGHGLEVEINCT | 120 |
| Patient_2 | KPCPPGERKARDCTVNGDEPDCVPCQEGKEYTDKAHFSSKCRRCRLCDEGHGLEVEINCT | 120 |
| Fas | RTQNTKCRCKPNFFCNSTVCEHCDPCTKCEHGIIKECTLTSNTKCKEEGSRSNLGWLCLL | 180 |
| Patient_1 | RTQNTKCRCKPNFFCNSTVCEHCDPCTKCEHGIIKECTLTSNTKCKEEGSRSNLGWLCLL | 180 |
| Patient_2 | RTQNTKCRCKPNFFCNSTVCEHCDPCTKCEHGIIKECTLTSNTKCKEEGSRSNLGWLCLL | 180 |
| Fas | LLPIPLIVWVKRKEVQKTCRKHRKENQGSHESPTLNPETVAINLSDVDLSKYITTIAGVM | 240 |
| Patient_1 | LLPIPLIVWVKRKEVQKTCRKHRKENQGSHESPTLNPETVAINLSDVDLSKYITTIAGVM | 240 |
| Patient_2 | LLPIPLIVWVKRKEVQKTCRKHRKENQGSHESPTLNPETVAINLSDVDLSKYITTIAGVM | 240 |
| Fas | TLSQVKGFVRKNGVNEAKIDEIKNDNVQDTAEQKVQLLRNWHQLHGKKEAYDTLIKDLKK | 300 |
| Patient_1 | TLSQVKGFVRKNGVNEAKIDEIKNDNVQDTAEQKVQLLRNWHQLHGKKEAYDTLIKDLKK | 300 |
| Patient_2 | TLSQVKGFVStop | 249 |
| Fas | ANLCTLAEKIQTIILKDITSDSENSNFRNEIQSLV | 335 |
| Patient_1 | ANLCTLAEKIQTIILKDITSDSENSNFRNEIQSLV | 335 |
| Patient_2 | 249 |
| Caspase_10 | MKSQGQHWYSSSDKNCKVSFREKLLIIDSNLGVQDVENLKFLCIGLVPNKKLEKSSSASD | 60 |
| Patient_1 | MKSQGQHWYSSSDKNCKVSFREKLLIIDSNLGVQDVENLKFLCIGLVPNKKLEKSSSASD | 60 |
| Patient_2 | MKSQGQHWYSSSDKNCKVSFREKLLIIDSNLGVQDVENLKFLCIGLVPNKKLEKSSSASD | 60 |
| Caspase_10 | VFEHLLAEDLLSEEDPFFLAELLYIIRQKKLLQHLNCTKEEVERLLPTRQRVSLFRNLLY | 120 |
| Patient_1 | VFEHLLAEDLLSEEDPFFLAELLYIIRQKKLLQHLNCTKEEVERLLPTRQRVSLFRNLLY | 120 |
| Patient_2 | VFEHLLAEDLLSEEDPFFLAELLYIIRQKKLLQHLNCTKEEVERLLPTRQRVSLFRNLLY | 120 |
| Caspase_10 | ELSEGIDSENLKDMIFLLKDSLPKTEMTSLSFLAFLEKQGKIDEDNLTCLEDLCKTVVPK | 180 |
| Patient_1 | ELSEGIDSENLKDMIFLLKDSLPKTEMTSLSFLAFLEKQGKIDEDNLTCLEDLCKTVVPK | 180 |
| Patient_2 | ELSEGIDSENLKDMIFLLKDSLPKTEMTSLSFLAFLEKQGKIDEDNLTCLEDLCKTVVPK | 180 |
| Caspase_10 | LLRNIEKYKREKAIQIVTPPVDKEAESYQGEEELVSQTDVKTFLEALPQESWQNKHAGSN | 240 |
| Patient_1 | LLRNIEKYKREKAIQIVTPPVDKEAESYQGEEELVSQTDVKTFLEALPQESWQNKHAGSN | 240 |
| Patient_2 | LLRNIEKYKREKAIQIVTPPVDKEAESYQGEEELVSQTDVKTFLEALPQESWQNKHAGSN | 240 |
| Caspase_10 | GNRATNGAPSLVSRGMQGASANTLNSETSTKRAAVYRMNRNHRGLCVIVNNHSFTSLKDR | 300 |
| Patient_1 | GNRATNGAPSLVSRGMQGASANTLNSETSTKRAAVYRMNRNHRGLCVIVNNHSFTSLKDR | 300 |
| Patient_2 | GNRATNGAPSLVSRGMQGASANTLNSETSTKRAAVYRMNRNHRGLCVIVNNHSFTSLKDR | 300 |
| * | ||
| Caspase_10 | QGTHKDAEILSHVFQWLGFTVHIHNNVTKVEMEMVLQKQKCNPAHADGDCFVFCILTHGR | 360 |
| Patient_1 | QGTHKDAEILSHVFQWLGFTVHIHNNVTKVEMEMVLQKQKCNPAHADGDCFVFCILTHGR | 360 |
| Patient_2 | QGTHKDAEILSHVFQWLGFTVHIHNNVTKVEMEMVLQKQKCNPAHADGDCFVFCILTHGR | 360 |
| Caspase_10 | FGAVYSSDEALIPIREIMSHFTALQCPRLAEKPKLFFIQACQGEEIQPSVSIEADALNPE | 420 |
| Patient_1 | FGAVYSSDEALIPIREIMSHFTALQCPRLAEKPKLFFIQACQGEEIQPSVSIEADALNPE | 420 |
| Patient_2 | FGAVYSSDEALIPIREIMSHFTALQCPRLAEKPKLFFIQACQGEEIQPSVSIEADALNPE | 420 |
| * | ||
| Caspase_10 | QAPTSLQDSIPAEADFLLGLATVPGYV | 447 |
| Patient_1 | QAPTSLQDSIPAEADFLLGLATVPGYV | 447 |
| Patient_2 | QAPTSLQDSIPAEADFLLGLATVPGYV | 447 |
| * |
| FasL | MQQPFNYPYPQIYWVDSSASSPWAPPGTVLPCPTSVPRRPGQRRPPPPPPPPPLPPPPPP | 60 |
| Patient_1 | MQQPFNYPYPQIYWVDSSASSPWAPPGTVLPCPTSVPRRPGQRRPPPPPPPPPLPPPPPP | 60 |
| Patient_2 | MQQPFNYPYPQIYWVDSSASSPWAPPGTVLPCPTSVPRRPGQRRPPPPPPPPPLPPPPPP | 60 |
| FasL | PPLPPLPLPPLKKRGNHSTGLCLLVMFFMVLVALVGLGLGMFQLFHLQKELAELRESTSQ | 120 |
| Patient_1 | PPLPPLPLPPLKKRGNHSTGLCLLVMFFMVLVALVGLGLGMFQLFHLQKELAELRESTSQ | 120 |
| Patient_2 | PPLPPLPLPPLKKRGNHSTGLCLLVMFFMVLVALVGLGLGMFQLFHLQKELAELRESTSQ | 120 |
| * * | ||
| FasL | MHTASSLEKQIGHPSPPPEKKELRKVAHLTGKSNSRSMPLEWEDTYGIVLLSGVKYKKGG | 180 |
| Patient_1 | MHTASSLEKQIGHPSPPPEKKELRKVAHLTGKSNSRSMPLEWEDTYGIVLLSGVKYKKGG | 180 |
| Patient_2 | MHTASSLEKQIGHPSPPPEKKELRKVAHLTGKSNSRSMPLEWEDTYGIVLLSGVKYKKGG | 180 |
| * | ||
| FasL | LVINETGLYFVYSKVYFRGQSCNNLPLSHKVYMRNSKYPQDLVMMEGKMMSYCTTGQMWA | 240 |
| Patient_1 | LVINETGLYFVYSKVYFRGQSCNNLPLSHKVYMRNSKYPQDLVMMEGKMMSYCTTGQMWA | 240 |
| Patient_2 | LVINETGLYFVYSKVYFRGQSCNNLPLSHKVYMRNSKYPQDLVMMEGKMMSYCTTGQMWA | 240 |
| FasL | RSSYLGAVFNLTSADHLYVNVSELSLVNFEESQTFFGLYKL | 281 |
| Patient_1 | RSSYLGAVFNLTSADHLYVNVSELSLVNFEESQTFFGLYKL | 281 |
| Patient_2 | RSSYLGAVFNLTSADHLYVNVSELSLVNFEESQTFFGLYKL | 281 |
| * * * |
| Primer | Sequence | Reference |
|---|---|---|
| Fas | ||
| FasA | 5′-AAGCTCTTTCACTTCGGAGG3′ | [37] |
| FasRL | 5′-CAATGTGTCATACGCTTCT-3′ | [37] |
| FasE | 5′-AGGACATGGCTTAGAAGTG-3′ | [37] |
| FasRN | 5′-ACAGCCAGCTATTAAGAAT-3′ | [37] |
| FasL | ||
| FasLs | 5′-TAAAACCGTTTGCTGGGGCT-3′ | This work |
| FasLas | 5′-TCGGAGTTCTGCCAGCTCCTT-3′ | This work |
| FasLasb | 5′-ATTGAACACTGCCCCCAGGT-3′ | This work |
| FasL423 | 5′-AAGGAGCTGGCAGAACTCCGA-3′ | This work |
| FasL757 | 5′AGGATCTGGTGATGATGGAG-3′ | This work |
| FasL3′ Del Rey | 5′-GAGAAGCACTTTGGGATTCTTTCC-3 | [21] |
| Casp10 | ||
| Ash | 5′-CCATGAAATCTCAAGGTCAACATTGG-3′ | [49] |
| WangV7R | 5′-GCATAGTCTTCAGGTGGGCGTTTG-3′ | [16] |
| 319F | 5′GTGGAGCGACTGCTGCCCACCCGA3′ | This work |
| 592F | ATCCAGATAGTGACACCTCCTGTA3′ | This work |
| e9bs | 5‘-CGAAAGTGGAAATGGAGATGGT-3′ | [50] |
| e9cas | 5′-CCACATGCCGAAAGGATACA-3′ | [50] |
| E2F7 | ||
| E2F7forClements | 5′-GGAAAGGCAACAGCAAACTCT-3′ | [51] |
| E2F7revClements | 5′-TGGGAGAGCACCAAGAGTAGAAGA-3′ | [51] |
| CRLF3 | ||
| For_CRLF3_Yang | 5′-AACGTTGATTACCAGTTCAG-3′ | [52] |
| Rev_CRLF3_Yang | 5′-CTGAGGACAGCTACGTTAGA-3′ | [52] |
| MLH3 | ||
| MLH3DMFor721 | 5′-GAGAAGGTTAGGCAGAGAATA-3′ | This work |
| RC1141 | 5′-ACAGAATTGGCACTGCACATT-3′ | This work |
| RPL30 F-RPL30 R-RPL30 | ||
| 5′-CTCCCAAAGGCTATTCAGTAATGG-3′ | Tapia-Ramírez J., personal communication | |
| 5′-GCTAAAAGGTGCTCGCTTCAGC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgadillo, D.M.; Céspedes-Cruz, A.I.; Ríos-Castro, E.; Rodríguez Maldonado, M.G.; López-Nogueda, M.; Márquez-Gutiérrez, M.; Villalobos-Manzo, R.; Ramírez-Reyes, L.; Domínguez-Fuentes, M.; Tapia-Ramírez, J. Differential Expression of Proteins in an Atypical Presentation of Autoimmune Lymphoproliferative Syndrome. Int. J. Mol. Sci. 2022, 23, 5366. https://doi.org/10.3390/ijms23105366
Delgadillo DM, Céspedes-Cruz AI, Ríos-Castro E, Rodríguez Maldonado MG, López-Nogueda M, Márquez-Gutiérrez M, Villalobos-Manzo R, Ramírez-Reyes L, Domínguez-Fuentes M, Tapia-Ramírez J. Differential Expression of Proteins in an Atypical Presentation of Autoimmune Lymphoproliferative Syndrome. International Journal of Molecular Sciences. 2022; 23(10):5366. https://doi.org/10.3390/ijms23105366
Chicago/Turabian StyleDelgadillo, Dulce María, Adriana Ivonne Céspedes-Cruz, Emmanuel Ríos-Castro, María Guadalupe Rodríguez Maldonado, Mariel López-Nogueda, Miguel Márquez-Gutiérrez, Rocío Villalobos-Manzo, Lorena Ramírez-Reyes, Misael Domínguez-Fuentes, and José Tapia-Ramírez. 2022. "Differential Expression of Proteins in an Atypical Presentation of Autoimmune Lymphoproliferative Syndrome" International Journal of Molecular Sciences 23, no. 10: 5366. https://doi.org/10.3390/ijms23105366
APA StyleDelgadillo, D. M., Céspedes-Cruz, A. I., Ríos-Castro, E., Rodríguez Maldonado, M. G., López-Nogueda, M., Márquez-Gutiérrez, M., Villalobos-Manzo, R., Ramírez-Reyes, L., Domínguez-Fuentes, M., & Tapia-Ramírez, J. (2022). Differential Expression of Proteins in an Atypical Presentation of Autoimmune Lymphoproliferative Syndrome. International Journal of Molecular Sciences, 23(10), 5366. https://doi.org/10.3390/ijms23105366