HOXA9 Overexpression Contributes to Stem Cell Overpopulation That Drives Development and Growth of Colorectal Cancer
Abstract
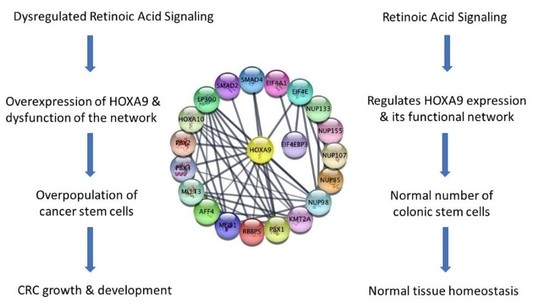
:1. Introduction
2. Results
2.1. Cell Proliferation Is Reduced by Modulating RA Signaling with ATRA and DEAB
2.2. ATRA and DEAB Reduce the Expression of Stem Cell Markers and Increase Cell Differentiation
2.3. ATRA and DEAB Reduce HOXA9 Expression
2.4. HOXA9 Is Selectively Expressed in Colonic SCs and Overexpressed in Human Colon Cancers
2.5. A Network of Proteins Associated with HOXA9 Is Classified as Being Transcriptionally Dysregulated in Cancer, and Genes Encoding Them Contain RAR and RXR Binding Sites
3. Discussion
4. Materials & Methods
4.1. Cell Culture
4.2. ATRA and DEAB Treatments
4.3. Cell Proliferation Assay
4.4. Western Blotting
4.5. Nanostring Profiling
4.6. Immunohistochemistry (IHC) and Immunofluorescence (IF) Microscopy
4.7. Bioinformatics
4.8. RNA Isolation, cDNA Conversion, and qPCR
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osmond, B.; Zhang, C.; Dinh, V.; Boman, B.M.; Facey, O.B. HOX Genes and Cancer Stem Cells: Update on HOX Expression, DNA Methylation, Biomarker Status, and Genetic Changes in Colorectal Cancer. J. Stem Cells Res. Dev. Ther. 2020, 6, 1–12. [Google Scholar] [CrossRef]
- Smith, J.; Zyoud, A.; Allegrucci, C. A Case of Identity: HOX Genes in Normal and Cancer Stem Cells. Cancers 2019, 11, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, A. Role of Hox genes in stem cell differentiation. World J. Stem Cells 2015, 7, 583. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018, 2018, 3569493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manfra, M.; Feliciello, I.; Bolognese, A.; Basilicata, M.G.; Pepe, G.; Campiglia, P.; Procino, A. Novel Anticancer Drug 5H-pyro[3,2-a] Phenoxazin-5-one (PPH) Regulates lncRNA HOTAIR and HOXC genes in Human MCF-7 Cells. Bahrain Med. Bull. 2021, 43, 334–341. [Google Scholar]
- Bhatlekar, S.; Viswanathan, V.; Fields, J.Z.; Boman, B.M. Overexpression of HOXA4 and HOXA9 genes promotes self-renewal and contributes to colon cancer stem cell overpopulation. J. Cell. Physiol. 2018, 233, 727–735. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Addya, S.; Salunek, M.; Orr, C.R.; Surrey, S.; McKenzie, S.; Fields, J.Z.; Boman, B.M. Identification of a Developmental Gene Expression Signature, Including HOX Genes, for the Normal Human Colonic Crypt Stem Cell Niche: Overexpression of the Signature Parallels Stem Cell Overpopulation during Colon Tumorigenesis. Stem Cells Dev. 2014, 23, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Bhatlekar, S.; Ertel, A.; Gonye, G.E.; Fields, J.Z.; Boman, B.M. Gene expression signatures for HOXA4, HOXA9, and HOXD10 reveal alterations in transcriptional regulatory networks in colon cancer. J. Cell. Physiol. 2019, 234, 13042–13056. [Google Scholar] [CrossRef]
- Aryal, S.; Zhang, Y.; Wren, S.; Li, C.; Lu, R. Molecular regulators of HOXA9 in acute myeloid leukemia. FEBS J. 2021. [Google Scholar] [CrossRef]
- Lambert, M.; Alioui, M.; Jambon, S.; Depauw, S.; Van Seuningen, I.; David-Cordonnier, M.-H. Direct and Indirect Targeting of HOXA9 Transcription Factor in Acute Myeloid Leukemia. Cancers 2019, 11, 837. [Google Scholar] [CrossRef] [Green Version]
- Depauw, S.; Lambert, M.; Jambon, S.; Paul, A.; Peixoto, P.; Nhili, R.; Marongiu, L.; Figeac, M.; Dassi, C.; Paul-Constant, C.; et al. Heterocyclic Diamidine DNA Ligands as HOXA9 Transcription Factor Inhibitors: Design, Molecular Evaluation, and Cellular Consequences in a HOXA9-Dependant Leukemia Cell Model. J. Med. Chem. 2019, 62, 1306–1329. [Google Scholar] [CrossRef]
- Watanabe, Y.; Saito, M.; Saito, K.; Matsumoto, Y.; Kanke, Y.; Onozawa, H.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; et al. Upregulated HOXA9 expression is associated with lymph node metastasis in colorectal cancer. Oncol. Lett. 2017, 15, 2756–2762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, T.; Jin, H.; Ying, R.; Xie, Q.; Zhou, C.; Gao, D. Overexpression of NAC1 confers drug resistance via HOXA9 in colorectal carcinoma cells. Mol. Med. Rep. 2017, 16, 3194–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janmaat, V.T.; Liu, H.; da Silva, R.A.; Wisse, P.H.A.; Spaander, M.C.W.; Ten Hagen, T.L.M.; Smits, R.; Bruno, M.J.; Fuhler, G.M.; Peppelenbosch, M.P. HOXA9 mediates and marks premalignant compartment size expansion in colonic adenomas. Carcinogenesis 2019, 40, 1514–1524. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. Klin. Wochenschr. 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Wang, X.; Bu, J.; Liu, X.; Wang, W.; Mai, W.; Lv, B.; Zou, J.; Mo, X.; Li, X.; Wang, J.; et al. miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer. Oncotarget 2017, 8, 63935–63948. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Zhang, Q.; Dong, M.; Yu, Y. MicroRNA-638 inhibits the progression of breast cancer through targeting HOXA9 and suppressing Wnt/β-cadherin pathway. World J. Surg. Oncol. 2021, 19, 247. [Google Scholar] [CrossRef]
- Pineault, N.; Helgason, C.D.; Lawrence, H.J.; Humphries, R.K. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 2002, 30, 49–57. [Google Scholar] [CrossRef]
- Facey, C.O.B.; Boman, B.M. Retinoids in Treatment of Colorectal Cancer. In Colorectal Cancer; BoD—Books on Demand: Norderstedt, Germany, 2020. [Google Scholar]
- Mongan, N.P.; Gudas, L.J. Diverse actions of retinoid receptors in cancer prevention and treatment. Differentiation 2007, 75, 853–870. [Google Scholar] [CrossRef]
- Nolte, C.; De Kumar, B.; Krumlauf, R. Hoxgenes: Downstream effectors of retinoic acid signaling in vertebrate embryogenesis. Genesis 2019, 57, e23306. [Google Scholar] [CrossRef]
- Modarai, S.R.; Gupta, A.; Opdenaker, L.M.; Kowash, R.; Masters, G.; Viswanathan, V.; Zhang, T.; Fields, J.Z.; Boman, B.M. The anti-cancer effect of retinoic acid signaling in CRC occurs via decreased growth of ALDH+ colon cancer stem cells and increased differentiation of stem cells. Oncotarget 2018, 9, 34658–34669. [Google Scholar] [CrossRef] [Green Version]
- Ginestier, C.; Wicinski, J.; Cervera, N.; Monville, F.; Finetti, P.; Bertucci, F.; Wicha, M.S.; Birnbaum, D.; Charafe-Jauffret, E. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle 2009, 8, 3297–3302. [Google Scholar] [CrossRef] [PubMed]
- Hunsu, V.O.; Facey, C.O.B.; Fields, J.Z.; Boman, B.M. Retinoids as Chemo-Preventive and Molecular-Targeted Anti-Cancer Therapies. Int. J. Mol. Sci. 2021, 22, 7731. [Google Scholar] [CrossRef] [PubMed]
- Opdenaker, L.M.; Kowash, R.; Masters, G.; Boman, B.M.; Zhang, T.; Modarai, S.R. Increased Musashi-2 and Decreased NUMB Protein Levels Observed in Human Colorectal Cancer are reverted to Normal Levels by ATRA-Induced Cell Differentiation. Int. J. Cancer Res. Ther. 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Croker, A.K.; Rodriguez-Torres, M.; Xia, Y.; Pardhan, S.; Leong, H.S.; Lewis, J.D.; Allan, A.L. Differential Functional Roles of ALDH1A1 and ALDH1A3 in Mediating Metastatic Behavior and Therapy Resistance of Human Breast Cancer Cells. Int. J. Mol. Sci. 2017, 18, 2039. [Google Scholar] [CrossRef]
- Croker, A.K.; Allan, A.L. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res. Treat. 2011, 133, 75–87. [Google Scholar] [CrossRef]
- Guo, W.-W.; Liu, W.-J. Expression of homeobox A9 in myeloid leukemia cell line HL-60 and effect of drugs on its expression. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2012, 20, 300–304. [Google Scholar]
- Mohr, S.; Doebele, C.; Comoglio, F.; Berg, T.; Beck, J.; Bohnenberger, H.; Alexe, G.; Corso, J.; Ströbel, P.; Wachter, A.; et al. Hoxa9 and Meis1 Cooperatively Induce Addiction to Syk Signaling by Suppressing miR-146a in Acute Myeloid Leukemia. Cancer Cell 2017, 31, 549–562.e11. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, M.W.; Durbin, A.D.; He, S.; Oppel, F.; Shi, H.; Tao, T.; Li, Z.; Berezovskaya, A.; Liu, Y.; Zhang, J.; et al. Retinoic acid rewires the adrenergic core regulatory circuitry of childhood neuroblastoma. Sci. Adv. 2021, 7, eabe0834. [Google Scholar] [CrossRef]
- Dihal, A.A.; Boot, A.; van Roon, E.H.; Schrumpf, M.; Fariña-Sarasqueta, A.; Fiocco, M.; Zeestraten, E.C.M.; Kuppen, P.J.K.; Morreau, H.; van Wezel, T.; et al. The Homeobox Gene MEIS1 Is Methylated in BRAFp.V600E Mutated Colon Tumors. PLoS ONE 2013, 8, e79898. [Google Scholar] [CrossRef] [Green Version]
- Dassé, E.; Volpe, G.; Walton, D.S.; Wilson, N.; Del Pozzo, W.; O’Neill, L.P.; Slany, R.K.; Frampton, J.; Dumon, S. Distinct regulation of c-myb gene expression by HoxA9, Meis1 and Pbx proteins in normal hematopoietic progenitors and transformed myeloid cells. Blood Cancer J. 2012, 2, e76. [Google Scholar] [CrossRef]
- Dobrotkova, V.; Chlapek, P.; Jezova, M.; Adamkova, K.; Mazanek, P.; Sterba, J.; Veselska, R. Prediction of neuroblastoma cell response to treatment with natural or synthetic retinoids using selected protein biomarkers. PLoS ONE 2019, 14, e0218269. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, H.; Hovland, R.; Forthun, R.B.; Erdal, S.; Gjertsen, B.T.; Fredly, H.; Bruserud, Ø. Disease-stabilizing treatment based on all-trans retinoic acid and valproic acid in acute myeloid leukemia—Identification of responders by gene expression profiling of pretreatment leukemic cells. BMC Cancer 2017, 17, 630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhao, C.; Zhao, J.; Zhu, Y.; Weng, X.; Chen, Q.; Sun, H.; Mi, J.-Q.; Li, J.; Zhu, J.; et al. Inactivation of PBX3 and HOXA9 by down-regulating H3K79 methylation represses NPM1-mutated leukemic cell survival. Theranostics 2018, 8, 4359–4371. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, X.; Escudero, R.; Farre, D.; Nunez, O.; Martinez, J.; Alba, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Farre, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Rosello, L.; Alba, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Ahn, K.; Emerick, B.; Modarai, S.R.; Opdenaker, L.M.; Palazzo, J.; Schleiniger, G.; Fields, J.Z.; Boman, B.M. APC mutations in human colon lead to decreased neuroendocrine maturation of ALDH+ stem cells that alters GLP-2 and SST feedback signaling: Clue to a link between WNT and retinoic acid signalling in colon cancer development. PLoS ONE 2020, 15, e0239601. [Google Scholar] [CrossRef]
- Kozovska, Z.; Patsalias, A.; Bajzik, V.; Durinikova, E.; Demkova, L.; Jargasova, S.; Smolkova, B.; Plava, J.; Kucerova, L.; Matuskova, M. ALDH1A inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer 2018, 18, 656. [Google Scholar] [CrossRef]






| Transcription Factor | Start Position * | End Position | TRE ** Sequence |
|---|---|---|---|
| RXR-alpha | −1454 | −1448 | TTTACCC |
| RXR-alpha | −1442 | −1436 | GGGTGAG |
| RXR-alpha | −1380 | −1374 | TTTACCC |
| RXR-alpha | −1168 | −1162 | GGGTCTC |
| RXR-alpha | −1021 | −1015 | GGGTCGC |
| RXR-alpha | −945 | −939 | GGGTCGA |
| RXR-alpha | −843 | −837 | GGGTCCT |
| RXR-alpha | −816 | −810 | GGGTTGC |
| RXR-alpha | −787 | −781 | GGGTAGC |
| RXR-alpha | −770 | −764 | TACACCC |
| RXR-alpha | −654 | −648 | GGGTAAA |
| RXR-alpha | −346 | −340 | GGGACCC |
| RXR-alpha | −205 | −199 | CAAACCC |
| PPAR-alpha:RXR-alpha | −1413 | −1403 | CAGGCCCAGGA |
| PPAR-alpha:RXR-alpha | −1025 | −1015 | GGCTGGGTCGC |
| RAR-beta | −817 | −808 | AGGGTTGCCC |
| RAR-beta | −207 | −198 | AACAAACCCC |
| RAR-beta:RXR-alpha | −42 | −31 | GGGCGCCGGCAA |
| Functional Classification | Genes |
|---|---|
| RNA transport | EIF4A1, EIF4EBP3, EIF4E, NUP107, NUP133, NUP155, NUP85, NUP98 |
| Transcriptional misregulation in cancer | MLLT3, MEIS1, PBX1, PBX3, HOXA10, KMT2A |
| Adherens junction | EP300, SMAD2, SMAD4 |
| TGF-beta signaling pathway | EP300, SMAD2, SMAD4 |
| Cell cycle | EP300, SMAD2, SMAD4 |
| FoxO signaling pathway | EP300, SMAD2, SMAD4 |
| Signaling pathways regulating pluripotency of stem cells | MEIS1, SMAD2, SMAD4 |
| Transcription Factor | MLLT3 | MEIS1 | HOXA10 | PBX1 | PBX3 | KMT2A | EIF4E | NUP98 | SMAD4 | PBX2 | NUP115 | NUP85 | NUP133 | NUP107 | EIF4EBP3 | EIF4A1 | RBBP5 | AFF4 | EP300 | SMAD2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RXRA | 11 | 15 | 21 | 4 | 8 | 13 | 8 | 15 | 7 | 18 | 11 | 11 | 8 | 5 | 10 | 12 | 9 | 14 | 15 | 13 |
| RARB | 2 | 4 | 7 | 1 | 1 | 6 | 1 | 4 | 2 | 4 | 4 | 5 | 4 | 2 | 4 | 3 | 1 | 4 | 4 | 3 |
| RARB: RXRA | 1 | 2 | 4 | 3 | 1 | 1 | 2 | 1 | 4 | 1 | 2 | 3 | 1 | 2 | 2 | |||||
| PXR-1/ RXRA | 3 | 2 | 3 | 2 | 3 | 2 | 1 | 3 | 1 | 3 | 2 | 3 | 4 | 1 | 3 | 3 | 1 | 3 | ||
| PPARA/ RXRA | 2 | 1 | 1 | 3 | 1 | 4 | 4 | 2 | 1 | 2 | 1 | 1 | 4 | 2 | 3 | 2 | ||||
| RARA1 | 1 | 2 | ||||||||||||||||||
| TOTAL | 17 | 24 | 35 | 8 | 13 | 25 | 14 | 29 | 15 | 31 | 18 | 21 | 15 | 13 | 25 | 16 | 16 | 26 | 20 | 23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osmond, B.; Facey, C.O.B.; Zhang, C.; Boman, B.M. HOXA9 Overexpression Contributes to Stem Cell Overpopulation That Drives Development and Growth of Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6799. https://doi.org/10.3390/ijms23126799
Osmond B, Facey COB, Zhang C, Boman BM. HOXA9 Overexpression Contributes to Stem Cell Overpopulation That Drives Development and Growth of Colorectal Cancer. International Journal of Molecular Sciences. 2022; 23(12):6799. https://doi.org/10.3390/ijms23126799
Chicago/Turabian StyleOsmond, Brian, Caroline O. B. Facey, Chi Zhang, and Bruce M. Boman. 2022. "HOXA9 Overexpression Contributes to Stem Cell Overpopulation That Drives Development and Growth of Colorectal Cancer" International Journal of Molecular Sciences 23, no. 12: 6799. https://doi.org/10.3390/ijms23126799
APA StyleOsmond, B., Facey, C. O. B., Zhang, C., & Boman, B. M. (2022). HOXA9 Overexpression Contributes to Stem Cell Overpopulation That Drives Development and Growth of Colorectal Cancer. International Journal of Molecular Sciences, 23(12), 6799. https://doi.org/10.3390/ijms23126799