Identification of Transcriptional Networks Involved in De Novo Organ Formation in Tomato Hypocotyl Explants
Abstract
:1. Introduction
2. Results
2.1. Expression of Genes Encoding TFs during De Novo Organ Formation
2.2. Analysis of TF Expression Profiles during De Novo Organ Formation
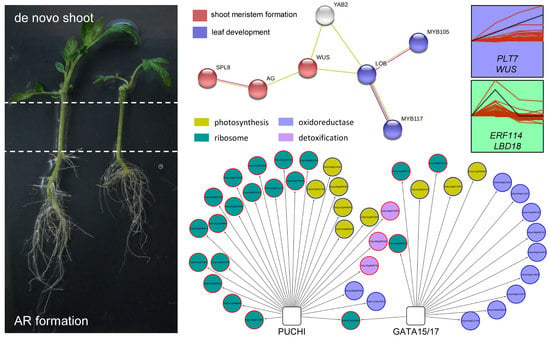
2.3. GRNs Required for De Novo Organ Formation
2.4. The Cell Cycle Is Differentially Regulated in the Apical and Basal Regions during de Novo Organ Formation
2.5. Characterization of ROS Accumulation during De Novo Organ Formation
2.6. CDF3 Is Required for De Novo Shoot Formation in Tomato Hypocotyl Explants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Tomato TF Identification and Annotation
4.3. RNA-Seq Analysis and GRN Generation
4.4. Cell Cycle Analysis
4.5. Cell Death and ROS Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant Regeneration: Cellular Origins and Molecular Mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular Mechanisms of Plant Regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, H. Genetic and Epigenetic Controls of Plant Regeneration. Curr. Top. Dev. Biol. 2014, 108, 1–33. [Google Scholar] [CrossRef]
- Mathew, M.M.; Prasad, K. Model Systems for Regeneration: Arabidopsis. Development 2021, 148, dev195347. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Shibata, M.; Rymen, B.; Iwase, A.; Bgman, A.M.; Watt, L.; Coleman, D.; Favero, D.S.; Takahashi, T.; Ahnert, S.E.; et al. A Gene Regulatory Network for Cellular Reprogramming in Plant Regeneration. Plant Cell Physiol. 2018, 59, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.H.; Noh, Y.S.; Seo, P.J. REGENOMICS: A Web-Based Application for Plant REGENeration-Associated TranscriptOMICS Analyses. Comput. Struct. Biotechnol. J. 2022, 20, 3234. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF Transcription Factor WIND1 Controls Cell Dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwase, A.; Mitsuda, N.; Ikeuchi, M.; Ohnuma, M.; Koizuka, C.; Kawamoto, K.; Imamura, J.; Ezura, H.; Sugimoto, K. Arabidopsis WIND1 Induces Callus Formation in Rapeseed, Tomato, and Tobacco. Plant Signal. Behav. 2013, 8, e27432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwase, A.; Mita, K.; Nonaka, S.; Ikeuchi, M.; Koizuka, C.; Ohnuma, M.; Ezura, H.; Imamura, J.; Sugimoto, K. WIND1-Based Acquisition of Regeneration Competency in Arabidopsis and Rapeseed. J. Plant Res. 2015, 128, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Iwase, A.; Harashima, H.; Ikeuchi, M.; Rymen, B.; Ohnuma, M.; Komaki, S.; Morohashi, K.; Kurata, T.; Nakata, M.; Ohme-Takagi, M.; et al. WIND1 Promotes Shoot Regeneration through Transcriptional Activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 2017, 29, 54–69. [Google Scholar] [CrossRef]
- Iwase, A.; Kondo, Y.; Laohavisit, A.; Takebayashi, A.; Ikeuchi, M.; Matsuoka, K.; Asahina, M.; Mitsuda, N.; Shirasu, K.; Fukuda, H.; et al. WIND Transcription Factors Orchestrate Wound-Induced Callus Formation, Vascular Reconnection and Defense Response in Arabidopsis. New Phytol. 2021, 232, 734–752. [Google Scholar] [CrossRef] [PubMed]
- Alaguero-Cordovilla, A.; Sánchez-García, A.B.; Ibáñez, S.; Albacete, A.; Cano, A.; Acosta, M.; Pérez-Pérez, J.M. An Auxin-Mediated Regulatory Framework for Wound-Induced Adventitious Root Formation in Tomato Shoot Explants. Plant Cell Environ. 2021, 44, 1642–1662. [Google Scholar] [CrossRef]
- Larriba, E.; Sánchez-García, A.B.; Martínez-Andújar, C.; Albacete, A.; Pérez-Pérez, J.M. Tissue-Specific Metabolic Reprogramming during Wound-Induced Organ Formation in Tomato Hypocotyl Explants. Int. J. Mol. Sci. 2021, 22, 10112. [Google Scholar] [CrossRef] [PubMed]
- Larriba, E.; Belén Sánchez-García, A.; Salud Justamante, M.; Martínez-Andújar, C.; Albacete, A.; Manuel Pérez-Pérez, J.; Tran, P.; Golam Mostofa, M. Dynamic Hormone Gradients Regulate Wound-Induced de Novo Organ Formation in Tomato Hypocotyl Explants. Int. J. Mol. Sci. 2021, 22, 11843. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. IDEP: An Integrated Web Application for Differential Expression and Pathway Analysis of RNA-Seq Data. BMC Bioinform. 2018, 19, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, J.; Bar-Joseph, Z. STEM: A Tool for the Analysis of Short Time Series Gene Expression Data. BMC Bioinform. 2006, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Omary, M.; Gil-Yarom, N.; Yahav, C.; Steiner, E.; Hendelman, A.; Efroni, I. A Conserved Superlocus Regulates Above- and Belowground Root Initiation. Science 2022, 375, eabf4368. [Google Scholar] [CrossRef]
- Vlieghe, K.; Boudolf, V.; Beemster, G.T.S.; Maes, S.; Magyar, Z.; Atanassova, A.; De Almeida Engler, J.; De Groodt, R.; Inzé, D.; De Veylder, L. The DP-E2F-like Gene DEL1 Controls the Endocycle in Arabidopsis Thaliana. Curr. Biol. 2005, 15, 59–63. [Google Scholar] [CrossRef]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral Root Initiation Requires the Sequential Induction of Transcription Factors LBD16 and PUCHI in Arabidopsis Thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef]
- Ranftl, Q.L.; Bastakis, E.; Klermund, C.; Schwechheimer, C. LLM-Domain Containing B-GATA Factors Control Different Aspects of Cytokinin-Regulated Development in Arabidopsis Thaliana. Plant Physiol. 2016, 170, 2295–2311. [Google Scholar] [CrossRef]
- Goh, T.; Joi, S.; Mimura, T.; Fukaki, H. The Establishment of Asymmetry in Arabidopsis Lateral Root Founder Cells Is Regulated by LBD16/ASL18 and Related LBD/ASL Proteins. Development 2012, 139, 883–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA Genes Mediate Patterning of the Arabidopsis Root Stem Cell Niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochatt, S.J. Medicago Truncatula Handbook Flow Cytometry (Ploidy Determination, Cell Cycle Analysis, DNA Content per Nucleus). 2006. Available online: https://www.researchgate.net/publication/342002480_Medicago_truncatula_protocols_flow_cytometry_ploidy_determination_cell_cycle_analysis_DNA_content_per_nucleus (accessed on 11 November 2022).
- Takahashi, I.; Kojima, S.; Sakaguchi, N.; Umeda-Hara, C.; Umeda, M. Two Arabidopsis Cyclin A3s Possess G1 Cyclin-like Features. Plant Cell Rep. 2010, 29, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Skylar, A.; Chang, P.L.; Bisova, K.; Wu, X. CYCP2;1 Integrates Genetic and Nutritional Information to Promote Meristem Cell Division in Arabidopsis. Dev. Biol. 2014, 393, 160–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A Database for ROS Homeostasis Regulated Proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Zandalinas, S.I.; Fichman, Y.; Mittler, R. Integration of Reactive Oxygen Species and Hormone Signaling during Abiotic Stress. Plant J. 2021, 105, 459–476. [Google Scholar] [CrossRef]
- Sagi, M.; Davydov, O.; Orazova, S.; Yesbergenova, Z.; Ophir, R.; Stratmann, J.W.; Fluhr, R. Plant Respiratory Burst Oxidase Homologs Impinge on Wound Responsiveness and Development in Lycopersicon Esculentum. Plant Cell 2004, 16, 616–628. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Renau-Morata, B.; Carrillo, L.; Dominguez-Figueroa, J.; Vicente-Carbajosa, J.; Molina, R.V.; Nebauer, S.G.; Medina, J.; Doerner, P. CDF Transcription Factors: Plant Regulators to Deal with Extreme Environmental Conditions. J. Exp. Bot. 2020, 71, 3803–3815. [Google Scholar] [CrossRef]
- Reinhardt, D.; Frenz, M.; Mandel, T.; Kuhlemeier, C. Microsurgical and Laser Ablation Analysis of Interactions between the Zones and Layers of the Tomato Shoot Apical Meristem. Development 2003, 130, 4073–4083. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David-Schwartz, R.; Koenig, D.; Sinha, N.R. LYRATE Is a Key Regulator of Leaflet Initiation and Lamina Outgrowth in Tomato. Plant Cell 2009, 21, 3093–3104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiessl, K.; Muiño, J.M.; Sablowski, R. Arabidopsis JAGGED Links Floral Organ Patterning to Tissue Growth by Repressing Kip-Related Cell Cycle Inhibitors. Proc. Natl. Acad. Sci. USA 2014, 111, 2830–2835. [Google Scholar] [CrossRef] [Green Version]
- Capua, Y.; Eshed, Y. Coordination of Auxin-Triggered Leaf Initiation by Tomato LEAFLESS. Proc. Natl. Acad. Sci. USA 2017, 114, 3246–3251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, Y.; Kawamura, A.; Suzuki, T.; Segami, S.; Maeshima, M.; Polyn, S.; De Veylder, L.; Sugimoto, K. Transcriptional Activation of Auxin Biosynthesis Drives Developmental Reprogramming of Differentiated Cells. Plant Cell 2022, 34, 4348–4365. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, N.; Wang, L.; Li, J.; Zheng, X.; Xiang, F.; Liu, Z. MYB94 and MYB96 Additively Inhibit Callus Formation via Directly Repressing LBD29 Expression in Arabidopsis Thaliana. Plant Sci. 2020, 293, 110323. [Google Scholar] [CrossRef]
- Domínguez-Figueroa, J.; Carrillo, L.; Renau-Morata, B.; Yang, L.; Molina, R.V.; Marino, D.; Canales, J.; Weih, M.; Vicente-Carbajosa, J.; Nebauer, S.G.; et al. The Arabidopsis Transcription Factor CDF3 Is Involved in Nitrogen Responses and Improves Nitrogen Use Efficiency in Tomato. Front. Plant Sci. 2020, 11, 1825. [Google Scholar] [CrossRef]
- Kareem, A.; Durgaprasad, K.; Sugimoto, K.; Du, Y.; Pulianmackal, A.J.; Trivedi, Z.B.; Abhayadev, P.V.; Pinon, V.; Meyerowitz, E.M.; Scheres, B.; et al. PLETHORA Genes Control Regeneration by a Two-Step Mechanism. Curr. Biol. 2015, 25, 1017–1030. [Google Scholar] [CrossRef] [Green Version]
- Hendelman, A.; Zebell, S.; Rodriguez-Leal, D.; Dukler, N.; Robitaille, G.; Wu, X.; Kostyun, J.; Tal, L.; Wang, P.; Bartlett, M.E.; et al. Conserved Pleiotropy of an Ancient Plant Homeobox Gene Uncovered by Cis-Regulatory Dissection. Cell 2021, 184, 1724–1739.e16. [Google Scholar] [CrossRef]
- Bellande, K.; Trinh, D.C.; Gonzalez, A.A.; Dubois, E.; Petitot, A.S.; Lucas, M.; Champion, A.; Gantet, P.; Laplaze, L.; Guyomarc’H, S. PUCHI Represses Early Meristem Formation in Developing Lateral Roots of Arabidopsis Thaliana. J. Exp. Bot. 2022, 73, 3496–3510. [Google Scholar] [CrossRef]
- Bolt, S.; Zuther, E.; Zintl, S.; Hincha, D.K.; Schmülling, T. ERF105 Is a Transcription Factor Gene of Arabidopsis Thaliana Required for Freezing Tolerance and Cold Acclimation. Plant Cell Environ. 2017, 40, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-Mediated Wound Signalling Promotes Plant Regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Hashimoto, N.; Kawai, S.; Yumoto, E.; Shibata, K.; Tameshige, T.; Yamamoto, Y.; Sugimoto, K.; Asahina, M.; Ikeuchi, M. Auxin-Induced WUSCHEL-RELATED HOMEOBOX13 Mediates Asymmetric Activity of Callus Formation upon Cutting. Plant Cell Physiol. 2022. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Ito, T.; Tanaka, H.; Favero, D.S.; Kawamura, A.; Sakamoto, S.; Wakazaki, M.; Tameshige, T.; Fujii, H.; et al. Wound-Inducible WUSCHEL-RELATED HOMEOBOX 13 Is Required for Callus Growth and Organ Reconnection. Plant Physiol. 2022, 188, 425–441. [Google Scholar] [CrossRef]
- Sakakibara, K.; Reisewitz, P.; Aoyama, T.; Friedrich, T.; Ando, S.; Sato, Y.; Tamada, Y.; Nishiyama, T.; Hiwatashi, Y.; Kurata, T.; et al. WOX13-like Genes Are Required for Reprogramming of Leaf and Protoplast Cells into Stem Cells in the Moss Physcomitrella Patens. Development 2014, 141, 1660–1670. [Google Scholar] [CrossRef] [Green Version]
- Asahina, M.; Azuma, K.; Pitaksaringkarn, W.; Yamazaki, T.; Mitsuda, N.; Ohme-Takagi, M.; Yamaguchi, S.; Kamiya, Y.; Okada, K.; Nishimura, T.; et al. Spatially Selective Hormonal Control of RAP2.6L and ANAC071 Transcription Factors Involved in Tissue Reunion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16128–16132. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, K.; Sato, R.; Matsukura, Y.; Kawajiri, Y.; Iino, H.; Nozawa, N.; Shibata, K.; Kondo, Y.; Satoh, S.; Asahina, M. Wound-Inducible ANAC071 and ANAC096 Transcription Factors Promote Cambial Cell Formation in Incised Arabidopsis Flowering Stems. Commun. Biol. 2021, 4, 1–12. [Google Scholar] [CrossRef]
- Vera-Sirera, F.; De Rybel, B.; Úrbez, C.; Kouklas, E.; Pesquera, M.; Álvarez-Mahecha, J.C.; Minguet, E.G.; Tuominen, H.; Carbonell, J.; Borst, J.W.; et al. A BHLH-Based Feedback Loop Restricts Vascular Cell Proliferation in Plants. Dev. Cell 2015, 35, 432–443. [Google Scholar] [CrossRef] [Green Version]
- De Rybel, B.; Möller, B.; Yoshida, S.; Grabowicz, I.; Barbier de Reuille, P.; Boeren, S.; Smith, R.S.; Borst, J.W.; Weijers, D. A BHLH Complex Controls Embryonic Vascular Tissue Establishment and Indeterminate Growth in Arabidopsis. Dev. Cell 2013, 24, 426–437. [Google Scholar] [CrossRef]
- Willems, A.; Heyman, J.; Eekhout, T.; Achon, I.; Pedroza-Garcia, J.A.; Zhu, T.; Li, L.; Vercauteren, I.; Van den Daele, H.; van de Cotte, B.; et al. The Cyclin CYCA3;4 Is a Postprophase Target of the APC/CCCS52A2 E3-Ligase Controlling Formative Cell Divisions in Arabidopsis. Plant Cell 2020, 32, 2979–2996. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Suzuki, T.; Iwata, E.; Nakamichi, N.; Suzuki, T.; Chen, P.; Ohtani, M.; Ishida, T.; Hosoya, H.; Müller, S.; et al. Transcriptional Repression by MYB3R Proteins Regulates Plant Organ Growth. EMBO J. 2015, 34, 1992–2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cejudo, F.J.; González, M.-C.; Pérez-Ruiz, J.M. Redox Regulation of Chloroplast Metabolism. Plant Physiol. 2020, 186, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Mhimdi, M.; Pérez-Pérez, J.M. Understanding of Adventitious Root Formation: What Can We Learn From Comparative Genetics? Front. Plant Sci. 2020, 11, 1541. [Google Scholar] [CrossRef] [PubMed]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional Regulation of ROS Controls Transition from Proliferation to Differentiation in the Root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Marcos, M.; Desvoyes, B.; Manzano, C.; Liberman, L.M.; Benfey, P.N.; del Pozo, J.C.; Gutierrez, C. Control of Arabidopsis Lateral Root Primordium Boundaries by MYB36. New Phytol. 2017, 213, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Minibayeva, F.; Beckett, R.P.; Kranner, I. Roles of Apoplastic Peroxidases in Plant Response to Wounding. Phytochemistry 2015, 112, 122–129. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse Functions and Reactions of Class III Peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-Wide Identification of Arabidopsis LBD29 Target Genes Reveals the Molecular Events behind Auxin-Induced Cell Reprogramming during Callus Formation. Plant Cell Physiol. 2018, 59, 749–760. [Google Scholar] [CrossRef]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN Transcription Factors Direct Callus Formation in Arabidopsis Regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, T.T.; Liu, H.; Shi, D.Y.; Wang, M.; Bie, X.M.; Li, X.G.; Zhang, X.S. Thioredoxin-Mediated ROS Homeostasis Explains Natural Variation in Plant Regeneration. Plant Physiol. 2018, 176, 2231–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, N.; Mittler, R. Reactive Oxygen Species-Dependent Wound Responses in Animals and Plants. Free Radic. Biol. Med. 2012, 53, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Nagao, A.; Kobayashi, M.; Koyasu, S.; Chow, C.C.T.; Harada, H. HIF-1-Dependent Reprogramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int. J. Mol. Sci. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, K.E.; Joy, S.; Delhove, J.M.K.M.; Kotiadis, V.N.; Fernandez, E.; Fitzpatrick, L.M.; Whiteford, J.R.; King, P.J.; Bolanos, J.P.; Duchen, M.R.; et al. NRF2 Orchestrates the Metabolic Shift during Induced Pluripotent Stem Cell Reprogramming. Cell Rep. 2016, 14, 1883–1891. [Google Scholar] [CrossRef] [Green Version]
- Pucciariello, C.; Perata, P. The Oxidative Paradox in Low Oxygen Stress in Plants. Antioxidants 2021, 10, 332. [Google Scholar] [CrossRef]
- Renau-Morata, B.; Molina, R.V.; Carrillo, L.; Cebolla-Cornejo, J.; Sánchez-Perales, M.; Pollmann, S.; Domínguez-Figueroa, J.; Corrales, A.R.; Flexas, J.; Vicente-Carbajosa, J.; et al. Ectopic Expression of CDF3 Genes in Tomato Enhances Biomass Production and Yield under Salinity Stress Conditions. Front. Plant Sci. 2017, 8, 660. [Google Scholar] [CrossRef] [Green Version]
- Feller, C.; Bleiholder, H.; Buhr, L.; Hack, H.; Hess, M.; Klose, R.; Meier, U.; Stauss, R.; Van den Boom, T.; Weber, E. Phänologische Entwicklungsstadien von Gemüsepflanzen: II. Fruchtgemüse Und Hülsenfrüchte. Nachrichtenbl. Deut. Pflanzenschutzd. 1995, 47, 217–232. [Google Scholar]
- Sol Genomics Network. Available online: https://solgenomics.net/organism/Solanum_lycopersicum/genome/ (accessed on 11 November 2022).
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. ITAK: A Program for Genome-Wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Sinharoy, S.; Udvardi, M.; Zhao, P.X. PlantTFcat: An Online Plant Transcription Factor and Transcriptional Regulator Categorization and Analysis Tool. BMC Bioinform. 2013, 14, 321. [Google Scholar] [CrossRef] [Green Version]
- Hmmscan. Available online: https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan (accessed on 11 November 2022).
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)Orthologs in Large-Scale Analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Araport 11 Genome Release. Available online: https://www.arabidopsis.org/download/ (accessed on 11 November 2022).
- Morpheus. Available online: https://software.broadinstitute.org/morpheus/ (accessed on 11 November 2022).
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting Functional Regulatory Maps in Plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Cytoscape. Available online: https://cytoscape.org/ (accessed on 11 November 2022).
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid Flow Cytometric Analysis of the Cell Cycle in Intact Plant Tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Ai, G.; Zhang, D.; Huang, R.; Zhang, S.; Li, W.; Ahiakpa, J.K.; Zhang, J. Genome-Wide Identification and Molecular Characterization of the Growth-Regulating Factors-Interacting Factor Gene Family in Tomato. Genes 2020, 11, 1435. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Gao, S.; Xiong, C.; Yu, G.; Chang, J.; Ye, Z.; Yang, C. Comprehensive analysis and expression profile of the homeodomain leucine zipper IV transcription factor family in tomato. Plant Physiol. Biochem. 2015, 96, 141–153. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Huang, Z.; Van Houten, J.; Gonzalez, G.; Xiao, H.; van der Knaap, E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013, 288, 111–129. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, L.; Zhang, Y.; Xu, L.; Li, N.; Zhang, X.; Pan, Y. Genome-wide identification of C2H2 zinc-finger genes and their expression patterns under heat stress in tomato (Solanum lycopersicum L.). PeerJ 2019, 7, e7929. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 288. [Google Scholar] [CrossRef] [PubMed]
- Khatun, K.; Robin, A.H.K.; Park, J.-I.; Nath, U.K.; Kim, C.K.; Lim, K.-B.; Nou, I.S.; Chung, M.-Y. Molecular Characterization and Expression Profiling of Tomato GRF Transcription Factor Family Genes in Response to Abiotic Stresses and Phytohormones. Int. J. Mol. Sci. 2017, 18, 1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Peng, R.; Tian, Y.; Han, H.; Xu, J.; Yao, Q. Genome-Wide Identification and Analysis of the MYB Transcription Factor Superfamily in Solanum lycopersicum. Plant Cell Physiol. 2016, 57, 1657–1677. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Hamyat, M.; Liu, C.; Ahmad, S.; Gao, X.; Guo, C.; Wang, Y.; Guo, Y. Identification and Characterization of the WOX Family Genes in Five Solanaceae Species Reveal Their Conserved Roles in Peptide Signaling. Genes 2018, 9, 260, Erratum in Genes 2018, 9, 457. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Zhao, T.; Xu, X.; Li, J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum). PeerJ 2017, 5, e3955. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hu, Z.; Zhao, T.; Yang, Y.; Chen, T.; Yang, M.; Yu, W.; Zhang, B. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genom. 2015, 16, 39. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef] [Green Version]
- Xu, R. Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 965–979. [Google Scholar] [CrossRef]
- Xing, H.; Jiang, Y.; Zou, Y.; Long, X.; Wu, X.; Ren, Y.; Li, Y.; Li, H.L. Genome-wide investigation of the AP2/ERF gene family in ginger: Evolution and expression profiling during development and abiotic stresses. BMC Plant Biol. 2021, 21, 561. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhang, C.; Zhao, T.; Yao, M.; Xu, X. A genome-wide analysis of GATA transcription factor family in tomato and analysis of expression patterns. Int. J. Agric. Biol. 2018, 20, 1274–1282. [Google Scholar]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Saegusa, M.; Iwamoto, K.; Oda, Y.; Katayama, H.; Kojima, M.; Sakakibara, H.; Fukuda, H. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 2014, 24, 2053–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliaga Fandino, A.C.; Kim, H.; Rademaker, J.D.; Lee, J.Y. Reprogramming of the cambium regulators during adventitious root development upon wounding of storage tap roots in radish (Raphanus sativus L.). Biol. Open 2019, 8, bio039677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnyk, C.W.; Gabel, A.; Hardcastle, T.J.; Robinson, S.; Miyashima, S.; Grosse, I.; Meyerowitz, E.M. Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E2447–E2456. [Google Scholar] [CrossRef] [Green Version]
- Han, G.; Wei, X.; Dong, X.; Wang, C.; Sui, N.; Guo, J.; Yuan, F.; Gong, Z.; Li, X.; Zhang, Y.; et al. Arabidopsis ZINC FINGER PROTEIN1 Acts Downstream of GL2 to Repress Root Hair Initiation and Elongation by Directly Suppressing bHLH Genes. Plant Cell 2020, 32, 206–225. [Google Scholar] [CrossRef] [Green Version]
- Chandler, J.W. Class VIIIb APETALA2 Ethylene Response Factors in Plant Development. Trends Plant Sci. 2018, 23, 151–162. [Google Scholar] [CrossRef]
- Sengupta, S.; Nag Chaudhuri, R. ABI3 plays a role in de-novo root regeneration from Arabidopsis thaliana callus cells. Plant Signal Behav. 2020, 15, 1794147. [Google Scholar] [CrossRef]
- Durgaprasad, K.; Roy, M.V.; Venugopal, M.A.; Kareem, A.; Raj, K.; Willemsen, V.; Mähönen, A.P.; Scheres, B.; Prasad, K. Gradient Expression of Transcription Factor Imposes a Boundary on Organ Regeneration Potential in Plants. Cell Rep. 2019, 29, 453–463.e3. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.P.; Zhou, C.; Wang, S.S.; Yuan, J.; Zhang, X.S.; Su, Y.H. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 2017, 213, 1740–1754. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Bergmann, D.C. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 2007, 134, 2959–2968. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Parra, E.; Perianez-Rodriguez, J.; Navarro-Neila, S.; Gude, I.; Moreno-Risueno, M.A.; Del Pozo, J.C. The transcription factor OBP4 controls root growth and promotes callus formation. New Phytol. 2017, 213, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Zhang, H.K.; Zhai, J.F.; Zhang, X.S.; Sang, Y.L.; Cheng, Z.J. ARF4 regulates shoot regeneration through coordination with ARF5 and IAA12. Plant Cell Rep. 2021, 40, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.B.; Shang, G.D.; Pan, Y.; Xu, Z.G.; Zhou, C.M.; Mao, Y.B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.W. AP2/ERF Transcription Factors Integrate Age and Wound Signals for Root Regeneration. Plant Cell. 2020, 32, 226–241. [Google Scholar] [CrossRef] [Green Version]
- Canher, B.; Lanssens, F.; Zhang, A.; Bisht, A.; Mazumdar, S.; Heyman, J.; Wolf, S.; Melnyk, C.W.; De Veylder, L. The regeneration factors ERF114 and ERF115 regulate auxin-mediated lateral root development in response to mechanical cues. Mol. Plant. 2022, 15, 1543–1557. [Google Scholar] [CrossRef]
- De Lucas, M.; Pu, L.; Turco, G.; Gaudinier, A.; Morao, A.K.; Harashima, H.; Kim, D.; Ron, M.; Sugimoto, K.; Roudier, F.; et al. Transcriptional Regulation of Arabidopsis Polycomb Repressive Complex 2 Coordinates Cell-Type Proliferation and Differentiation. Plant Cell. 2016, 28, 2616–2631. [Google Scholar] [CrossRef] [Green Version]
- Efroni, I.; Mello, A.; Nawy, T.; Ip, P.L.; Rahni, R.; DelRose, N.; Powers, A.; Satija, R.; Birnbaum, K.D. Root Regeneration Triggers an Embryo-like Sequence Guided by Hormonal Interactions. Cell 2016, 165, 1721–1733. [Google Scholar] [CrossRef] [Green Version]
- Heyman, J.; Cools, T.; Canher, B.; Shavialenka, S.; Traas, J.; Vercauteren, I.; Van den Daele, H.; Persiau, G.; De Jaeger, G.; Sugimoto, K.; et al. The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat. Plants 2016, 2, 16165. [Google Scholar] [CrossRef]
- Sabatini, S.; Heidstra, R.; Wildwater, M.; Scheres, B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003, 17, 354–358. [Google Scholar] [CrossRef] [Green Version]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Paul, P.; Hartman, J.M.; Perry, S.E. AGL15 Promotion of Somatic Embryogenesis: Role and Molecular Mechanism. Front. Plant Sci. 2022, 13, 861556. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Verstraeten, I.; Werbrouck, S.; Geelen, D. CUC2 as an early marker for regeneration competence in Arabidopsis root explants. J. Plant Physiol. 2011, 168, 1598–1601. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, A.T. Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Choi, M.H.; Noh, B.; Noh, Y.S. De Novo Shoot Regeneration Controlled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 2020, 61, 1600–1613. [Google Scholar] [CrossRef]
- Zhai, N.; Xu, L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants 2021, 7, 1453–1460. [Google Scholar] [CrossRef]
- Hassani, S.B.; Trontin, J.F.; Raschke, J.; Zoglauer, K.; Rupps, A. Constitutive Overexpression of a Conifer WOX2 Homolog Affects Somatic Embryo Development in Pinus pinaster and Promotes Somatic Embryogenesis and Organogenesis in Arabidopsis Seedlings. Front. Plant Sci. 2022, 13, 838421. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larriba, E.; Nicolás-Albujer, M.; Sánchez-García, A.B.; Pérez-Pérez, J.M. Identification of Transcriptional Networks Involved in De Novo Organ Formation in Tomato Hypocotyl Explants. Int. J. Mol. Sci. 2022, 23, 16112. https://doi.org/10.3390/ijms232416112
Larriba E, Nicolás-Albujer M, Sánchez-García AB, Pérez-Pérez JM. Identification of Transcriptional Networks Involved in De Novo Organ Formation in Tomato Hypocotyl Explants. International Journal of Molecular Sciences. 2022; 23(24):16112. https://doi.org/10.3390/ijms232416112
Chicago/Turabian StyleLarriba, Eduardo, Míriam Nicolás-Albujer, Ana Belén Sánchez-García, and José Manuel Pérez-Pérez. 2022. "Identification of Transcriptional Networks Involved in De Novo Organ Formation in Tomato Hypocotyl Explants" International Journal of Molecular Sciences 23, no. 24: 16112. https://doi.org/10.3390/ijms232416112
APA StyleLarriba, E., Nicolás-Albujer, M., Sánchez-García, A. B., & Pérez-Pérez, J. M. (2022). Identification of Transcriptional Networks Involved in De Novo Organ Formation in Tomato Hypocotyl Explants. International Journal of Molecular Sciences, 23(24), 16112. https://doi.org/10.3390/ijms232416112