Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract
Abstract
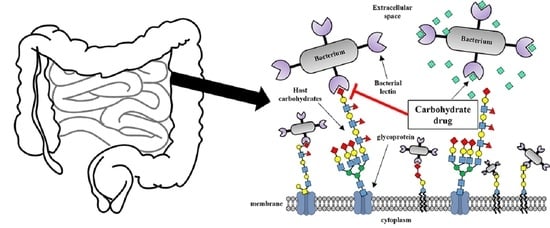
:1. Introduction
2. Lectin-Glycan Interactions
2.1. Ecosystems in the Human Gastrointestinal Tract (GIT) in Relation to LGI
2.2. Immune Responses in the GIT of Hosts in Connection with LGI
2.3. Outline and Present Status of LGI Research
2.4. Strategies to Search for Lectins of Pathogenic Intestinal Bacteria
3. Conclusions
Future Perspectives of LGI Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, C.-H. Glycobiology in Innate Immunology; Nature Springer Publishing, Co.: Singapore, 2022. [Google Scholar]
- Winzler, R.J. Carbohydrates in Cell Surfaces. Int. Rev. Cytol. 1970, 29, 77–125. [Google Scholar] [PubMed]
- McCoy, J.P., Jr.; Chambers, W.H. Carbohydrates in the Functions of Natural Killer Cells. Glycobiology 1991, 1, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Gabius, H.J. Cell Surface Glycans: The Why and How of their Functionality as Biochemical Signals in Lectin-Mediated Information Transfer. Crit. Rev. Immunol. 2006, 26, 43–79. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-H. Ganglioside Biochemistry; Nature Springer Publishing, Co.: Singapore, 2020. [Google Scholar]
- Kim, C.-H. GM3 Signaling; Nature Springer Publishing, Co.: Singapore, 2021. [Google Scholar]
- Kim, C.-H. Glycosphingolipids Signaling; Nature Springer Publishing, Co.: Singapore, 2021. [Google Scholar]
- Choi, M.J.; Kim, Y.R.; Park, N.G.; Kim, C.H.; Oh, Y.D.; Lim, H.K.; Kim, J.M. Characterization of a C-Type Lectin Domain-Containing Protein with Antibacterial Activity from Pacific Abalone (Haliotis discus hannai). Int. J. Molec. Sci. 2022, 23, 698. [Google Scholar] [CrossRef] [PubMed]
- Suhas, B.; Sashikala, R.I. An Overview of Lectin-Glycan Interactions: A Key Event in Initiating Fungal Infection and Pathogenesis. Arch. Microbiol. 2018, 200, 371–382. [Google Scholar]
- Narla, S.N.; Sun, X.L. Glyco-Macroligand Microarray with Controlled Orientation and Glycan Density. Lab. Chip. 2012, 12, 1656–1663. [Google Scholar] [CrossRef]
- Kilpatrick, D.C. Lectin-Glycoconjugate Interactions in Health and Disease. Biochem. Soc. Trans. 2008, 36, 1453–1456. [Google Scholar] [CrossRef] [Green Version]
- Hart, D.A. Lectins in Biological Systems: Applications to Microbiology. Am. J. Clin. Nutr. 1980, 33, 2416–2425. [Google Scholar] [CrossRef] [Green Version]
- Mandal, C.; Mandal, C. Sialic Acid Binding Lectins. Experientia 1990, 46, 433–441. [Google Scholar] [CrossRef]
- Sharon, N. Lectins: Carbohydrate-Specific Reagents and Biological Recognition Molecules. J. Biol. Chem. 2007, 282, 2753–2764. [Google Scholar] [CrossRef] [Green Version]
- Wander, V.B.; Stefan, P.; Herman, W.F.; Raoul, J.G.; Hans, J.N. Bitter-Sweet Symphony: Glycan-Lectin Interactions in Virus Biology. FEMS Microbiol. Rev. 2014, 38, 598–632. [Google Scholar]
- Yu, H.; Chen, X. Carbohydrate Post-Glycosylational Modifications. Org. Biomol. Chem. 2007, 5, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Barroca, N.; Jacquinet, J.C. An Access to Various Sulfation Patterns in Dermatan Sulfate: Chemical Syntheses of Sulfoforms of Trisaccharide Methyl Glycosides. Carbohydr. Res. 2002, 337, 673–689. [Google Scholar] [CrossRef]
- Ha, S.H.; Kwak, C.H.; Park, J.Y.; Abekura, F.; Lee, Y.C.; Kim, J.S.; Chung, T.W.; Kim, C.H. 3′-Sialyllactose Targets Cell Surface Protein, SIGLEC-3, and Induces Megakaryocyte Differentiation and Apoptosis by Lipid Raft-Dependent Endocytosis. Glycoconj. J. 2020, 37, 187–200. [Google Scholar] [CrossRef]
- Cho, S.H.; Lee, K.M.; Kim, C.H.; Kim, S.S. Construction of a Lectin-Glycan Interaction Network from Enterohemorrhagic Escherichia Coli Strains by Multi-Omics Analysis. Int. J. Mol. Sci. 2020, 21, 2681. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the Mucosal Barrier to Infection. Mucosal. Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Wales, A.D.; Woodward, M.J.; Pearson, G.R. Attaching-Effacing Bacteria in Animals. J. Comp. Pathol. 2005, 132, 1–26. [Google Scholar] [CrossRef]
- Rogers, E.A.; Das, A.; Ton-That, H. Adhesion by Pathogenic Corynebacteria. Adv. Exp. Med. Biol. 2011, 715, 91–103. [Google Scholar]
- Amieva, M.R.; El-Omar, E.M. Host-Bacterial Interactions in Helicobacter Pylori Infection. Gastroenterology 2008, 134, 306–323. [Google Scholar] [CrossRef] [Green Version]
- Cummings, R.D. The Repertoire of Glycan Determinants in the Human Glycome. Mol. Biosyst. 2009, 5, 1087–1104. [Google Scholar] [CrossRef]
- Schnaar, R.L. The Biology of Gangliosides. Adv Carbohydr Chem. Biochem. 2019, 76, 113–148. [Google Scholar] [PubMed]
- Nakano, T.; Sugawara, M.; Kawakami, H. Sialic Acid in Human Milk: Composition and Functions. Acta Paediatr. Taiwan 2001, 42, 11–17. [Google Scholar]
- Varki, A. Multiple Changes in Sialic Acid Biology during Human Evolution. Glycoconj. J. 2009, 26, 231–245. [Google Scholar] [CrossRef]
- Wang, B.; Brand-Miller, J. The Role and Potential of Sialic Acid in Human Nutrition. Eur. J. Clin. Nutr. 2003, 57, 1351–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikulak, J.; Di Vito, C.; Zaghi, E.; Mavilio, D. Host Immune Responses in HIV-1 Infection: The Emerging Pathogenic Role of Siglecs and Their Clinical Correlates. Front. Immunol. 2017, 8, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledeen, R.W.; Kopitz, J.; Abad-Rodríguez, J.; Gabius, H.-J. Glycan Chains of Gangliosides: Functional Ligands for Tissue Lectins (Siglecs/Galectins). Prog. Mol. Biol. Transl. Sci. 2018, 156, 289–324. [Google Scholar] [PubMed]
- Iwabuchi, K.; Nakayama, H.; Oizumi, A.; Suga, Y.; Ogawa, H.; Takamori, K. Role of Ceramide from Glycosphingolipids and Its Metabolites in Immunological and Inflammatory Responses in Humans. Mediat. Inflamm. 2015, 2015, 120748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savage, P.B.; Teyton, L.; Bendelac, A. Glycolipids for Natural Killer T Cells. Chem. Soc. Rev. 2006, 35, 771–779. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Nizet, V. The Interplay between Siglecs and Sialylated Pathogens. Glycobiology 2014, 24, 818–825. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Nizet, V. Siglecs at the Host-Pathogen Interface. Adv. Exp. Med. Biol. 2020, 1204, 197–214. [Google Scholar]
- Jones, C.; Virji, M.; Crocker, P.R. Recognition of Sialylated Meningococcal Lipopolysaccharide by Siglecs Expressed on Myeloid Cells Leads to Enhanced Bacterial Uptake. Mol. Microbiol. 2003, 49, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Bochner, B.S.; Alvarez, R.A.; Mehta, P.; Bovin, N.V.; Blixt, O.; White, J.R.; Schnaar, R.L. Glycan Array Screening Reveals a Candidate Ligand for Siglec. J. Biol. Chem. 2005, 280, 4307–4312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkataraman, M.; Sasisekharan, R.; Raman, R. Glycan Array Data Management at Consortium for Functional Glycomics. Methods Mol. Biol. 2015, 1273, 181–190. [Google Scholar] [PubMed]
- Peng, W.; Nycholat, C.M.; Razi, N. Glycan Microarray Screening Assay for Glycosyltransferase Specificities. Methods Mol. Biol. 2013, 1022, 1–14. [Google Scholar] [PubMed]
- Avril, T.; Wagner, E.R.; Willison, H.J.; Crocker, P.R. “Sialic Acid-Binding Immunoglobulin-Like Lectin 7 Mediates Selective Recognition of Sialylated Glycans Expressed on Campylobacter Iejuni Lipooligosaccharides”. Infect. Immun. 2006, 74, 4133–4141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their Roles in the Immune System. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Varki, A.; Angata, T. Siglecs–the Major Subfamily of I-Type Lectins. Glycobiology 2006, 16, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Crocker, P.R. Evolution of CD33-Related Siglecs: Regulating Host Immune Functions and Escaping Pathogen Exploitation? Immunology 2011, 132, 18–26. [Google Scholar] [CrossRef]
- Ducreux, J.; Crocker, P.R.; Vanbever, R. Analysis of Sialoadhesin Expression on Mouse Alveolar Macrophages. Immunol. Lett. 2009, 124, 77–80. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Li, R.; Qiao, S.L.; Chen, X.X.; Deng, R.G.; Zhang, G.P. Porcine Sialoadhesin Suppresses Type I Interferon Production to Support Porcine Reproductive and Respiratory Syndrome Virus Infection. Vet. Res. 2020, 51, 18. [Google Scholar] [CrossRef] [Green Version]
- Dragacevic, L.; Djordjevic, B.; Gavrovic-Jankulovic, M.; Ilic, V.; Kanazir, D.; Minic, R. ELLSA Based Profiling of Surface Glycosylation in Microorganisms Reveals that ss-Glucan Rich Yeasts’ Surfaces are Selectively Recognized with Recombinant Banana Lectin. Glycoconj. J. 2020, 37, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Saijo, S.; Iwakura, Y. Dectin-1 and Dectin-2 in Innate Immunity against Fungi. Int. Immunol. 2011, 23, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Lahm, H.; André, S.; Hoeflich, A.; Kaltner, H.; Siebert, H.C.; Sordat, B.; von der Lieth, C.W.; Wolf, E.; Gabius, H.J. Tumor Galectinology: Insights into the Complex Network of a Family of Endogenous Lectins. Glycoconj. J. 2004, 20, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.K.; Syed, P.; Lehtinen, L.; Leivo, J.; Gidwani, K.; Wittfooth, S.; Pettersson, K.; Lamminmäki, U. A Nanoparticle-Based Approach for the Detection of Extracellular Vesicles. Sci. Rep. 2019, 11, 10038. [Google Scholar] [CrossRef]
- De Oliveira, D.M.; Hartley-Tassell, L.; Everest-Dass, A.; Day, C.J.; Dabbs, R.A.; Ve, T.; Kobe, B.; Nizet, V.; Packer, N.H.; Walker, M.J.; et al. Blood Group Antigen Recognition via the Group a Streptococcal M Protein Mediates Host Colonization. MBio 2017, 8, e02237-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossez, Y.; Gosset, P.; Boneca, I.G.; Magalhães, A.; Ecobichon, C.; Reis, C.A.; Cieniewski-Bernard, C.; Joncquel Chevalier Curt, M.; Léonard, R.; Maes, E.; et al. The lacdiNAc-Specific Adhesin LabA Mediates Adhesion of Helicobacter Pylori to Human Gastric Mucosa. J. Infect. Dis. 2014, 210, 1286–1295. [Google Scholar] [CrossRef] [Green Version]
- Sokurenko, E.V.; Chesnokova, V.; Dykhuizen, D.E.; Ofek, I.; Wu, X.R.; Krogfelt, K.A.; Struve, C.; Schembri, M.A.; Hasty, D.L. Pathogenic Adaptation of Escherichia Coli by Natural Variation of the FimH Adhesin. Proc. Natl Acad. Sci. USA 1998, 95, 8922–8926. [Google Scholar] [CrossRef] [Green Version]
- Spaulding, C.N.; Klein, R.D.; Ruer, S.; Kau, A.L.; Schreiber, H.L.; Cusumano, Z.T.; Dodson, K.W.; Pinkner, J.S.; Fremont, D.H.; Janetka, J.W.; et al. Selective Depletion of Uropathogenic E. Coli from the Gut by a FimH Antagonist. Nature 2017, 546, 528–532. [Google Scholar] [CrossRef] [Green Version]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. MBio 2016, 7, e00584-16. [Google Scholar] [CrossRef] [Green Version]
- Poole, J.; Day, C.J.; Itzstein, M.; Paton, J.C.; Jennings, M.P. Glycointeractions in Bacterial Pathogenesis. Nat. Rev. Microbiol. 2018, 16, 440–452. [Google Scholar]
- Lee, K.M.; Kim, C.H.; Kim, J.H.; Kim, S.S.; Cho, S.H. e-Membranome: A Database for Genome-Wide Analysis of Escherichia Coli Outer Membrane Proteins. Curr. Pharm. Biotechnol. 2021, 22, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Lee, K.M.; Kim, C.H.E. Coli Outer Membrane Protein Prediction Method and System for Performing the Method. KR Patent 10-2242972, 21 April 2021. [Google Scholar]
- Matiethoz, J.; Khatib, K.; Alocci, D.; Campbell, M.P.; Karlsson, N.G.; Packer, N.H.; Mullen, E.H.; Lisacek, F. SugarBindDB, a Resource of Glycan Mediated Host-Pathogen Interactions. Nucleic Acids Res. 2015, 44, 1243–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakhsheer, B.; Anderson, M.; Khatib, K.; Tadoori, L.; Joshi, L.; Lisacek, F.; Hirschman, L.; Mullen, E. SugarBind Database (SugarBindDB): A Resource of Pathogen Lectins and Corresponding Glycan Targets. J. Mol. Recognit. 2013, 26, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Bojar, D.; Powers, R.K.; Camacho, D.M.; Collins, J.J. Deep-Learning Resources for Studying Glycan-Mediated Host-Microbe Interactions. Cell Host Microbe. 2021, 29, 132–144. [Google Scholar] [CrossRef] [PubMed]
- SIB Swiss Institute of Bioinformatics Members. The SIB Swiss Institute of Bioinformatics’ Resources: Focus on Curated Databases. Nucleic Acids Res. 2016, 44, D27–D37. [Google Scholar] [CrossRef] [PubMed]
- Kubilis, A.; Abdulkarim, A.; Eissa, A.M.; Cameron, N.R. Giant Polymersome Protocells Dock with Virus Particle Mimics via Multivalent Glycan-Lectin Interactions. Sci. Rep. 2016, 6, 32414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pol-Fachin, L. Insights into the Effects of Glycosylation and the Monosaccharide-Binding Activity of the Plant Lectin CrataBL. Glycoconj. J. 2017, 34, 515–522. [Google Scholar] [CrossRef]
- Kerzmann, A.; Fuhrmann, J.; Kohlbacher, O.; Neumann, D. BALLDock/SLICK: A New Method for Protein-Carbohydrate Docking. J. Chem. Inf. Model 2008, 48, 1616–1625. [Google Scholar] [CrossRef]
- Boittier, E.D.; Burns, J.M.; Gandhi, N.S.; Ferro, V. GlycoTorch Vina: Docking Designed and Tested for Glycosaminoglycans. J. Chem. Inf. Model 2020, 60, 6328–6343. [Google Scholar] [CrossRef]
- Rosenberg, A.; Griffin, K.; Studier, F.W.; McCormick, M.; Berg, J.; Novy, R.; Mierendorf, R. T7Select Phage Display Systems: A Powerful New Protein Display System Based on Bacteriophage T7. Innovations 1996, 6, 1–6. [Google Scholar]
- Pulzova, L.; Flachbartova, Z.; Bencurova, E.; Potocnakova, L.; Comor, L.; Schreterova, E.; Bhide, M. Identification of B-Cell Epitopes of Borrelia Burgdorferi Outer Surface Protein C by Screening a Phage-Displayed Gene Fragment Library. Microbiol. Immunol. 2016, 60, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.J.; Volk, A.L.; Persson, H.; Säll, A.; Borrebaeck, C.; Uhlen, M.; Rockberg, J. Combination of Phage and Gram-Positive Bacterial Display of Human Antibody Repertoires Enables Isolation of Functional High Affinity Binders. New Biotechnol. 2018, 45, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Galán, A.; Comor, L.; Horvatić, A.; Kuleš, J.; Guillemin, N.; Mrljak, V.; Bhide, M. Library-Based Display Technologies: Where Do We Stand? Mol. Biosyst. 2016, 12, 2342–2358. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Troy, E.B.; Hu, L.T.; Gao, L.; Norris, S.J. Transposon Mutagenesis as an Approach to Improved Understanding of Borrelia Pathogenesis and Biology. Front. Cell Infect. Microbiol. 2014, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Veeranagouda, Y.; Husain, F.; Wexler, H.M. Transposon Mutagenesis of Bacteroides fragilis. Methods Mol. Biol. 2019, 2016, 105–116. [Google Scholar]
- Siegrist, M.S.; Rubin, E.J. Phage Transposon Mutagenesis. Methods Mol. Biol. 2009, 465, 311–323. [Google Scholar]
- Woodworth, D.L.; Kreuzer, K.N. A System of Transposon Mutagenesis for Bacteriophage T4. Mol. Microbiol. 1992, 6, 1289–1296. [Google Scholar] [CrossRef]
- Winterberg, K.M.; Reznikoff, W.S. Screening Transposon Mutant Libraries Using Full-Genome Oligonucleotide Microarrays. Methods Enzymol. 2007, 421, 110–125. [Google Scholar]
- Schüler, V.; Lussi, A.; Kage, A.; Seemann, R. Glycan-Binding Specificities of Streptococcus Mutans and Streptococcus Sobrinus Lectin-Like Adhesins. Clin. Oral Investig. 2012, 16, 789–796. [Google Scholar] [CrossRef]
- Lei, Y.; Yu, H.; Dong, Y.; Yang, J.; Ye, W.; Wang, Y.; Chen, W.; Jia, Z.; Xu, Z.; Li, Z.; et al. Characterization of N-Glycan Structures on the Surface of Mature Dengue 2 Virus Derived from Insect Cells. PLoS ONE 2015, 10, e0132122. [Google Scholar] [CrossRef] [Green Version]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; et al. Printed Covalent Glycan Array for Ligand Profiling of Diverse Glycan Binding Proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipke, P.N. Glycomics for Microbes and Microbiologists. MBio 2016, 7, e01224-16. [Google Scholar] [CrossRef] [Green Version]
- Kannappan, R.; Ando, M.; Furuhata, K.; Uda, Y. Photoaffinity Labeling of Sialidase with a Biotin-Conjugated Phenylaminodiazirine Derivative of 2,3-Didehydro-2-Deoxy-N-Acetylneuraminic Acid. Biol. Pharm. Bull. 2008, 31, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Li, P.J.; Anwar, M.T.; Fan, C.Y.; Juang, D.S.; Lin, H.Y.; Chang, T.C.; Kawade, S.K.; Chen, H.J.; Chen, Y.J.; Tan, K.T.; et al. Fluorescence “Turn-on” Lectin Sensors Fabricated by Ligand-Assisted Labeling Probes for Detecting Protein-Glycoprotein Interactions. Biomacromolecules 2020, 21, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Brzezicka, K.A.; Serna, S.; Reichardt, N.C. Fluorescent Neoglycoprotein Gold Nanoclusters: Synthesis and Applications in Plant Lectin Sensing and Cell Imaging. Nanoscale Res. Lett. 2018, 13, 360. [Google Scholar] [CrossRef] [PubMed]






| Lectin | Bacteria | Glycan |
|---|---|---|
| Shiga toxin | EHEC | Globotriaosylceramide (Gb3) |
| FedF | EHEC | H type 1, A6 type 1, H5 type 1, A6 type 1, Antigen H |
| E. coli K99 fimbriae | ETEC | NeuGc-GM3 (GM3gc) |
| CS3 | ETEC | GalNac (β1-4) Gal |
| 20 K fimbriae (CS31A surface adhesin) | ETEC | N-acetylglucosamine (GlcNAc) |
| S-fimbrial adhesin | ETEC | Siaα2-3Gal |
| Fimbrial adhesins | ETEC | Mannose (Man) |
| Bacterial adhesins | ETEC | Galabiose (Ga2), Siaα2-3Gal |
| Bacterial adhesin F17-G | ETEC | N-acetylglucosamine (GlcNAc) |
| Fimbriae Recognizing Sialyl Galactosides | ETEC | Siaα2-3Gal |
| FimH | ETEC | Mannose (Man) |
| LT | ETEC | NeuAc-GM1, NeuAc-GD1b, Asialo-GM1a |
| F4 fimbriae | ETEC | Lactosylceramide (LacCer), Globotriaosylceramide (Gb3), Hexaosylceramide, sulfatide, Asialo-GM2, Asialo-GM1, Galablosylceramide |
| Lectin | Bacteria | Glycan |
|---|---|---|
| Peb3 | Campylobacter jejuni | phosphoenolpyruvate, 3-phosphoglycerate |
| BC2L-C Nter | Burkholderia cenocepacia | H type 1 and Leb |
| BC2L-C Cter | Burkholderia cenocepacia | α-Man and a-mannoheptulose |
| Hif fimbriae | Haemophilus influenzae | α-Neup5Ac-(2-6)-β-Galp-(1-4)-GlcpNAc |
| CupB6 | Pseudomonas aeruginosa | Leb |
| LecA/PA-IL | Pseudomonas aeruginosa | α-Galp-(1-3)-β-Galp-(1-4) |
| LecB/PA-IIL | Pseudomonas aeruginosa | α-Fucp-(1-3/4)-β-GlcpNAc |
| BabA | Helicobacter pylori | Leb, A/H type 1, Globo A/H |
| SabA | Helicobacter pylori | α-Neup5Ac-(2-3)-β-Galp-(1-4)-b-GlcpNAc |
| FaeG | Escherichia coli | α-GalpNAc-(1-3)-β-GalpNAc-(1-3)-β-Galp-(1-4)- β-Glcp-(1-O)-Cer |
| FedF | Escherichia coli | ABH type 1 (Figure 2), sulphated H type 2 |
| PapG | Escherichia coli | β-GalpNAc-(1-3)-a-Galp-(1-4)- β-Glcp-(1-O)-Cer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, S.-H.; Park, J.-y.; Kim, C.-H. Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract. Int. J. Mol. Sci. 2022, 23, 1451. https://doi.org/10.3390/ijms23031451
Cho S-H, Park J-y, Kim C-H. Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract. International Journal of Molecular Sciences. 2022; 23(3):1451. https://doi.org/10.3390/ijms23031451
Chicago/Turabian StyleCho, Seung-Hak, Jun-young Park, and Cheorl-Ho Kim. 2022. "Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract" International Journal of Molecular Sciences 23, no. 3: 1451. https://doi.org/10.3390/ijms23031451
APA StyleCho, S. -H., Park, J. -y., & Kim, C. -H. (2022). Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract. International Journal of Molecular Sciences, 23(3), 1451. https://doi.org/10.3390/ijms23031451