Poly-L-Lysine–Lactobionic Acid-Capped Selenium Nanoparticles for Liver-Targeted Gene Delivery
Abstract
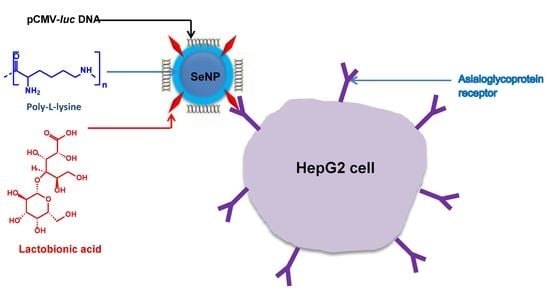
:1. Introduction
2. Results
2.1. Nanoparticle Synthesis and Characterization
2.2. Electrophoretic Mobility Shift Assay
2.3. Dye Displacement Assay
2.4. Enzyme Protection Assay
2.5. MTT Cell Viability Assay
2.6. Apoptosis
2.7. Reporter Gene and Receptor Competition Assay
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of Selenium Nanoparticles (SeNPs)
4.3. Preparation of Poly-L-Lysine Encapsulated SeNPs (PLL–SeNP)
4.4. Preparation of Lactobionic Acid-Modified PLL–SeNPs (LA–PLL–SeNP)
4.5. Characterization
4.5.1. UV-Visible Spectroscopy
4.5.2. Fourier-Transform Infrared Spectroscopy (FTIR)
4.5.3. Nanoparticle Tracking Analysis (NTA)
4.5.4. Transmission Electron Microscopy (TEM)
4.6. Electrophoretic Mobility Shift Assay
4.7. Dye Displacement Assay
4.8. Enzyme Protection Assay
4.9. MTT Cell Viability Assay
4.10. Luciferase Expression Assay
4.11. Competition Binding Assay
4.12. Apoptosis
4.13. Statistical Calculations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AO | Acridine orange |
| ASGP-R | Asialoorosomucoid receptor |
| BCA | Bicinchoninic acid |
| DMSO | Dimethyl sulfoxide |
| EB | Ethidium bromide |
| EDC | N-ethyl-N′-(3-dimethyl aminopropyl) carbodiimide |
| EDTA | Ethylenediamine tetraacetic acid |
| EMEM | Eagle’s Minimum Essential Medium |
| FBS | Fetal bovine serum |
| FLC | Fibrolamellar HCC |
| FSeNPs | Functionalized SeNPs |
| FTIR | Fourier-transform infrared spectroscopy |
| HBS | HEPES buffered saline |
| HCC | Hepatocellular carcinoma |
| HCC-CCA | Mixed hepatocellular cholangiocarcinoma |
| HEPES | 2-(4-(2-hydroxyethyl)-1-piperazinyl) ethane sulphonic acid |
| iCCA | Intrahepatic cholangiocarcinoma |
| KCl | Potassium chloride |
| LA | Lactobionic acid |
| LA–PLL–SeNP | Lactobionic acid-modified Poly-L-lysine encapsulated SeNPs |
| mRNA | Messenger RNA |
| MTT | (3-(4,5dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NHS | N-hydroxysuccinimide |
| NPs | Nanoparticles |
| NTA | Nanoparticle tracking analysis |
| PBS | Phosphate-buffered saline |
| PDI | Polydispersity index |
| pDNA | Plasmid DNA (Specifically referring to pCMV-Luc DNA in this study). |
| PLL | Poly-L-lysine |
| PLL–SeNP | Poly-L-lysine encapsulated SeNPs |
| RLU | Relative light units |
| RME | Receptor-mediated endocytosis |
| SDS | Sodium dodecyl sulphate |
| Se | Selenium |
| SeNPs | Selenium nanoparticles |
| SPR | Surface plasmon resonance |
| TBE | Tris-Borate-EDTA |
| TEM | Transmission electron microscopy |
| UV-vis | UV-visible spectroscopy |
References
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Sia, D.; Villanueva, A.; Friedman, S.L.; Llovet, J.M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 2017, 152, 745–761. [Google Scholar] [CrossRef] [Green Version]
- Ozakyol, A. Global epidemiology of hepatocellular carcinoma (HCC epidemiology). J. Gastrointest. Cancer 2017, 48, 238–240. [Google Scholar] [CrossRef]
- Philips, C.A.; Rajesh, S.; Nair, D.C.; Ahamed, R.; Abduljaleel, J.K.; Augustine, P. Hepatocellular Carcinoma in 2021: An Exhaustive Update. Cureus 2021, 13, e19274. [Google Scholar] [CrossRef]
- Nahon, P.; Zucman-Rossi, J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J. Hepatol. 2012, 57, 663–674. [Google Scholar] [CrossRef] [Green Version]
- Bruix, J.; Gores, G.J.; Mazzaferro, V. Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut 2014, 63, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Beg, S.; Alharbi, K.S.; Alruwali, N.B.; Alotaibi, N.H.; Almalki, W.H.; Alenzi, S.K.; Altowayan, W.M.; Alshammari, M.S.; Rahman, M. Nanotherapeutic systems for delivering cancer vaccines: Recent advances. Nanomedicine 2020, 15, 1527–1537. [Google Scholar] [CrossRef] [PubMed]
- Eunus, S.A.; Sharker, S.; Islam, M.T.; Khan, I.N.; Shaw, S.; Rahman, A.; Uddin, S.J.; Shill, M.C.; Rehman, S.; Das, N.; et al. Targeting cancer cells with nanotherapeutics and nanodiagnostics: Current status and future perspectives. Semin. Cancer Biol. 2021, 69, 52–68. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Quezada, S.A.; Larkin, J.; Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Kang, G.; Wang, T.; Huang, H. Tumor angiogenesis and anti-angiogenic gene therapy for cancer. Oncol. Lett. 2018, 16, 687–702. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.P.; Zeng, Q.H.; Lu, G.Q.; Yu, A.B. Inorganic nanoparticles as carriers for efficient cellular delivery. Chem. Eng. Sci. 2006, 61, 1027–1040. [Google Scholar] [CrossRef]
- Erathodiyil, N.; Ying, J.Y. Functionalization of inorganic nanoparticles for bioimaging applications. Acc. Chem. Res. 2011, 44, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-G. Preface. Curr. Drug Del. 2021, 18, 2–3. [Google Scholar] [CrossRef]
- Yu, D.-G.; Wang, M.; Ge, R. Strategies for sustained drug release from electrospun multi-layer nanostructures. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, e1772. [Google Scholar] [CrossRef]
- Li, D.; Wang, M.; Song, W.; Yu, D.-G.; Bligh, S.W.A. Electrospun Janus Beads-On-A-String Structures for Different Types of Controlled Release Profiles of Double Drugs. Biomolecules 2021, 11, 635. [Google Scholar] [CrossRef] [PubMed]
- Jagaran, K.; Singh, M. Nanomedicine for Neurodegenerative Disorders: Focus on Alzheimer’s and Parkinson’s Diseases. Int. J. Mol. Sci. 2021, 22, 9082. [Google Scholar] [CrossRef]
- Maiyo, F.; Singh, M. Polymerized selenium nanoparticles for folate-receptor-targeted delivery of Anti-Luc-siRNA: Potential for gene silencing. Biomedicines 2020, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Ferro, C.; Florindo, H.F.; Santos, H.A. Selenium Nanoparticles for Biomedical Applications: From Development and Characterization to Therapeutics. Adv. Healthc. Mater. 2021, 10, 2100598. [Google Scholar] [CrossRef]
- Benko, I.; Nagy, G.; Tanczos, B.; Ungvari, E.; Sztrik, A.; Eszenyi, P.; Prokisch, J.; Banfalvi, G. Subacute toxicity of nano-selenium compared to other selenium species in mice. Environ. Toxicol. Chem. 2012, 31, 2812–2820. [Google Scholar] [CrossRef]
- Forootanfar, H.; Adeli-Sardou, M.; Nikkhoo, M.; Mehrabani, M.; Amir-Heidari, B.; Shahverdi, A.R.; Shakibaie, M. Antioxidant and cytotoxic effect of biologically synthesized selenium nanoparticles in comparison to selenium dioxide. J. Trace Elem. Med. Biol. 2014, 28, 75–79. [Google Scholar] [CrossRef]
- Nayak, V.; Singh, K.R.B.; Singh, A.K.; Singh, R.P. Potentialities of selenium nanoparticles in biomedical science. N. J. Chem. 2021, 45, 2849–2878. [Google Scholar] [CrossRef]
- Maiyo, F.; Singh, M. Selenium Nanoparticles: Potential in Cancer Gene and Drug Delivery. Nanomedicine 2017, 12, 1075–1089. [Google Scholar] [CrossRef]
- Shakibaie, M.; Khorramizadeh, M.R.; Faramarzi, M.A.; Sabzevari, O.; Shahverdi, A.R. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase-2 expression. Biotechnol. Appl. Biochem. 2010, 56, 7–15. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.; Yu, H. Elemental selenium at nano size possesses lower toxicity without compromising the fundamental effect on selenoenzymes: Comparison with selenomethionine in mice. Free Radic. Biol. Med. 2007, 42, 1524–1533. [Google Scholar] [CrossRef]
- Hu, C.H.; Li, Y.L.; Xiong, L.; Zhang, H.M.; Xia, M.S. Comparative effects of nano elemental selenium and sodium selenite on selenium retention in broiler chickens. Anim. Feed Sci. Technol. 2012, 177, 204–210. [Google Scholar] [CrossRef]
- Rzigalinski, B.A.; Meehan, K.; Davis, R.M.; Xu, Y.; Miles, W.C.; Cohen, C.A. Radical nanomedicine. Nanomedicine 2006, 1, 399–412. [Google Scholar] [CrossRef]
- Shi, L.; Xun, W.; Yue, W.; Zhang, C.; Ren, Y.; Shi, L.; Wang, Q.; Yang, R.; Lei, F. Effect of sodium selenite, Se-yeast and nano-elemental selenium on growth performance, Se concentration and antioxidant status in growing male goats. Small Rumin. Res. 2011, 96, 49–52. [Google Scholar] [CrossRef]
- Singh, D.; Singh, M. Hepatocellular-targeted mRNA delivery using functionalized selenium nanoparticles in vitro. Pharmaceutics 2021, 13, 298. [Google Scholar] [CrossRef] [PubMed]
- Khurana, A.; Tekula, S.; Saifi, M.A.; Venkatesh, P.; Godugu, C. Therapeutic applications of selenium nanoparticles. Biomed. Pharmacother. 2019, 111, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kim, S.O.; Linardy, E.; Dreaden, E.C.; Zhdanov, V.P.; Hammond, P.T.; Cho, N.J. Influence of pH and surface chemistry on poly (L-lysine) adsorption onto solid supports investigated by quartz crystal microbalance with dissipation monitoring. J. Phys. Chem. 2015, 119, 10554–10565. [Google Scholar] [CrossRef]
- Ma, Y.H.; Peng, H.Y.; Yang, R.X.; Fang, N. Preparation of Lysine-Coated Magnetic Fe2O3 Nanoparticles and Influence on Viability of A549 Lung Cancer Cells. Asian Pac. J. Cancer Prev. 2014, 15, 8981–8985. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Su, J.; Zhao, Z.; Zheng, W.; Wu, H.; Zhang, Y.; Chen, T. Differential effects of amino acid surface decoration on the anticancer efficacy of selenium nanoparticles. Dalton Trans. 2014, 43, 1854–1861. [Google Scholar] [CrossRef]
- Danhier, F.; Feron, O.; Préat, V. To exploit the tumor microenvironment: Passive and active tumor targeting of nanocarriers for anticancer drug delivery. J. Control Release 2010, 148, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, J.; Ling, D. Surface engineering of nanoparticles for targeted delivery to hepatocellular carcinoma. Small 2018, 14, 1702037. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Ariatti, M. Targeted gene delivery into HepG2 cells using complexes containing DNA, cationized asialoorosomucoid and activated cationic liposomes. J. Control Release 2003, 92, 383–394. [Google Scholar] [CrossRef]
- Akinyelu, A.; Oladimeji, O.; Singh, M. Lactobionic Acid-Chitosan Functionalized Gold Coated Poly(lactide-co-glycolide) Nanoparticles for Hepatocyte Targeted Gene Delivery. Adv. Nat. Sci. Nanosci. Nanotechnol. 2020, 11, 045017. [Google Scholar] [CrossRef]
- Selmani, A.; Ulm, L.; Kasemets, K.; Kurvet, I.; Erceg, I.; Babir, R.; Pem, B.; Santini, P.; Marion, I.D.; Vinković, T.; et al. Stability and toxicity of differently coated selenium nanoparticles under model environmental exposure settings. Chemosphere 2020, 250, 126265. [Google Scholar] [CrossRef]
- Beekes, M.; Lasch, P.; Naumann, D. Analytical applications of Fourier transform-infrared (FT-IR) spectroscopy in microbiology and prion research. Vet. Microbiol. 2007, 123, 305–319. [Google Scholar] [CrossRef] [Green Version]
- García, M.A. Surface plasmons in metallic nanoparticles: Fundamentals and applications. J. Phys. D. 2011, 44, 283001. [Google Scholar] [CrossRef]
- Tam, S.K.; Dusseault, J.; Polizu, S.; Ménard, M.; Hallé, J.P.; Yahia, L. Physicochemical model of alginate–poly-l-lysine microcapsules defined at the micrometric/nanometric scale using ATR-FTIR, XPS, and ToF-SIMS. Biomater. Sci. 2005, 26, 6950–6961. [Google Scholar] [CrossRef]
- Pillay, N.S.; Daniels, A.; Singh, M. Folate-Targeted Transgenic Activity of Dendrimer Functionalized Selenium Nanoparticles In Vitro. Int. J. Mol. Sci. 2020, 21, 7177. [Google Scholar] [CrossRef]
- Honary, S.; Zahir, F. Effect of Zeta Potential on the Properties of Nano-Drug Delivery Systems-A Review (Part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. [Google Scholar] [CrossRef]
- Raviña, M.; Cubillo, E.; Olmeda, D.; Novoa-Carballal, R.; Fernandez-Megia, E.; Riguera, R.; Sánchez, A.; Cano, A.; Alonso, M.J. Hyaluronic acid/chitosan-g-poly (ethylene glycol) nanoparticles for gene therapy: An application for pDNA and siRNA delivery. Pharm. Res. 2010, 27, 2544–2555. [Google Scholar] [CrossRef] [Green Version]
- Bai, K.; Hong, B.; He, J.; Hong, Z.; Tan, R. Preparation and antioxidant properties of selenium nanoparticles-loaded chitosan microspheres. Int. J. Nanomed. 2017, 12, 4527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Ahn, R.; Van Den Bossche, J.; Thompson, D.; O’Halloran, T. Folate-mediated intracellular drug delivery increases the anticancer efficacy of nanoparticulate formulation of arsenic trioxide. Mol. Cancer. Ther. 2009, 8, 1955–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhotra, S.; Jha, N.; Desai, K. A superficial synthesis of selenium nanospheres using wet chemical approach. Int. J. Nanotechnol. Appl. 2014, 3, 7–14. [Google Scholar]
- Higuchi, Y.; Kawakami, S.; Fumoto, S.; Yamashita, F.; Hashida, M. Effect of the particle size of galactosylated lipoplex on hepatocyte-selective gene transfection after intraportal administration. Biol. Pharm. Bull. 2006, 29, 1521–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fumoto, S.; Nakadori, F.; Kawakami, S.; Nishikawa, M.; Yamashita, F.; Hashida, M. Analysis of hepatic disposition of galactosylated cationic liposome/plasmid DNA complexes in perfused rat liver. Pharm. Res. 2003, 20, 1452–1459. [Google Scholar] [CrossRef]
- Singh, M.; Rogers, B.C.; Ariatti, M. Targeting of glycosylated lipoplexes in HepG2 cells: Anomeric and C-4 epimeric preference of the asialoglycoprotein receptor. S. Afr. J. Sci. 2007, 103, 204–210. [Google Scholar]
- Ostolska, I.; Wisniewska, M. Application of the zeta potential measurements to explanation of colloidal Cr2O3 stability mechanism in the presence of the ionic polyamino acids. Colloid. Polym. Sci. 2014, 292, 2453–2464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prazeres, D.M.; Ferreira, G.N.; Monteiro, G.A.; Cooney, C.L.; Cabral, J.M. Large-scale production of pharmaceutical-grade plasmid DNA for gene therapy: Problems and bottlenecks. Trends Biotechnol. 1999, 17, 169–174. [Google Scholar] [CrossRef]
- Guo, Y.; Ma, Y.; Xu, L.; Li, J.; Yang, W. Conformational change induced reversible assembly/disassembly of poly-L-lysine-functionalized gold nanoparticles. J. Phys. Chem. 2007, 111, 9172–9176. [Google Scholar] [CrossRef]
- Estevez, H.; Garcia-Lidon, J.C.; Luque-Garcia, J.L.; Camara, C. Effects of chitosan-stabilized selenium nanoparticles on cell proliferation, apoptosis and cell cycle pattern in HepG2 cells: Comparison with other selenospecies. Colloids Surf. B. 2014, 122, 184–193. [Google Scholar] [CrossRef]
- Akinyelu, J.; Singh, M. Chitosan stabilized Gold-Folate-Poly(lactide-co-glycolide) Nanoplexes Facilitate Efficient Gene Delivery in Hepatic and Breast Cancer Cells. J. Nanosci. Nanotechnol. 2018, 18, 4478–4486. [Google Scholar] [CrossRef] [PubMed]
- Singh, M. Assessing Nucleic acid: Cationic Nanoparticle Interaction for Gene Delivery. In Bio-Carrier Vectors; Kumaran, N., Ed.; Springer-Nature: New York, NY, USA, 2021; Volume 2211, pp. 43–55. [Google Scholar] [CrossRef]
- Almutary, A.; Sanderson, B.J.S. The MTT and crystal violet assays: Potential confounders in nanoparticle toxicity testing. Int. J. Toxicol. 2016, 35, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Alam, J.; Cook, J.L. Reporter genes: Application to the study of mammalian gene transcription. Anal. Biochem. 1990, 188, 245–254. [Google Scholar] [CrossRef]
- Ramamoorth, M.; Narvekar, A. Non-viral vectors in gene therapy—An overview. J. Clin. Diagnostic Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef]
- Oberdörster, G.; Oberdörster, E.; Oberdörster, J. Nanotoxicology: An emerging, discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005, 113, 823–839. [Google Scholar] [CrossRef]
- Chauhan, P.; Chaudhary, S. Role of surface modification on selenium nanoparticles: Enumerating the optical, thermal and structural properties. Opt. Mater. 2019, 97, 109380. [Google Scholar] [CrossRef]
- Schwartz, A.L.; Genze, H.J.; Lodish, H.F. Recycling of the asialoglycoprotein receptor: Biochemical and immunocytochemical evidence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982, 300, 229–235. [Google Scholar] [CrossRef]
- Wu, G.Y.; Wu, C.H. Receptor-mediated in vitro gene transformation by a soluble DNA carrier system. J. Biol. Chem. 1987, 262, 4429–4432. [Google Scholar] [CrossRef]
- Maiyo, F.; Singh, M. Folate-Targeted mRNA Delivery Using Chitosan Functionalized Selenium Nanoparticles: Potential in Cancer Immunotherapy. Pharmaceuticals 2019, 12, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiyo, F.C.; Mbatha, L.S.; Singh, M. Selenium Nanoparticles in Folate-Targeted delivery of the pCMV-Luc DNA Reporter Gene. Curr. Nanosci. 2021, 17, 871–880. [Google Scholar] [CrossRef]
- Vrandečić, K.; Ćosić, J.; Ilić, J.; Ravnjak, B.; Selmani, A.; Galić, E.; Pem, B.; Barbir, R.; Vrček, I.V.; Vinković, T. Antifungal activities of silver 46- and selenium nanoparticles stabilized with different surface coating agents. Pest Manag. Sci. 2020, 76, 2021–2029. [Google Scholar] [CrossRef]
- Liu, S.X.; Xia, Z.S.; Zhong, Y.Q. Gene therapy in pancreatic cancer. World J. Gastroenterol. 2014, 20, 13343. [Google Scholar] [CrossRef] [PubMed]








| Nanoparticles | Nanocomplexes | ||||||
|---|---|---|---|---|---|---|---|
| Size (nm) | ζ Potential (mV) | PDI | pDNA:NP Ratio (w/w) | Size (nm) | ζ Potential (mV) | PDI | |
| SeNP | 75.7 ± 0.8 | −12.1 ± 0.2 | 0.00011 | - | - | - | - |
| PLL–SeNP | 84.7 ± 10 | 28.6 ± 10 | 0.014 | 1:2.7 | 118.7 ± 16.3 | −26.9 ± 0.6 | 0.0189 |
| LA–PLL–SeNP | 124.3 ± 3.2 | 25.0 ± 6.3 | 0.00066 | 1:14 | 164.5 ± 77 | −21.1 ± 0.3 | 0.0219 |
| Nanocomplex | Suboptimal Ratio (w/w) | Optimal Ratio (w/w) | Supraoptimal Ratio (w/w) |
|---|---|---|---|
| PLL–SeNP:pDNA | 1:2.2 | 1:2.7 | 1:3.2 |
| LA–PLL–SeNP:pDNA | 1:10.9 | 1:14 | 1:16 |
| Cell Line | Apoptotic Index | |
|---|---|---|
| PLL–SeNP | LA–PLL–SeNP | |
| HEK293 | 0.04 | 0.09 |
| HeLa | 0.08 | 0.07 |
| HepG2 | 0.13 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naidoo, S.; Daniels, A.; Habib, S.; Singh, M. Poly-L-Lysine–Lactobionic Acid-Capped Selenium Nanoparticles for Liver-Targeted Gene Delivery. Int. J. Mol. Sci. 2022, 23, 1492. https://doi.org/10.3390/ijms23031492
Naidoo S, Daniels A, Habib S, Singh M. Poly-L-Lysine–Lactobionic Acid-Capped Selenium Nanoparticles for Liver-Targeted Gene Delivery. International Journal of Molecular Sciences. 2022; 23(3):1492. https://doi.org/10.3390/ijms23031492
Chicago/Turabian StyleNaidoo, Shaigan, Aliscia Daniels, Saffiya Habib, and Moganavelli Singh. 2022. "Poly-L-Lysine–Lactobionic Acid-Capped Selenium Nanoparticles for Liver-Targeted Gene Delivery" International Journal of Molecular Sciences 23, no. 3: 1492. https://doi.org/10.3390/ijms23031492
APA StyleNaidoo, S., Daniels, A., Habib, S., & Singh, M. (2022). Poly-L-Lysine–Lactobionic Acid-Capped Selenium Nanoparticles for Liver-Targeted Gene Delivery. International Journal of Molecular Sciences, 23(3), 1492. https://doi.org/10.3390/ijms23031492