Molecular Beacon for Detection miRNA-21 as a Biomarker of Lung Cancer
Abstract
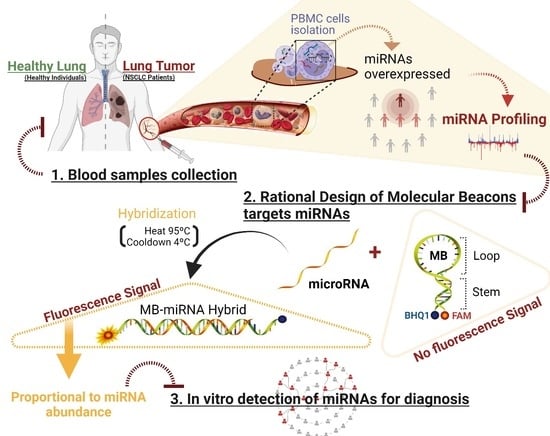
:1. Introduction
2. Results
2.1. miRNA Profiling
2.2. MB Design and Biophysical Characterization
2.3. Using the MB for Specific miRNA-21-5p Detection in Clinical Samples
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. RNA Isolation and Quantitative RT-PCR
4.3. MB Design and Synthetic Sequence Preparation
4.4. CD Spectroscopy
4.5. Fluorescence Resonance Energy Transfer (FRET) Melting
4.6. Nuclear Magnetic Resonance Spectroscopy
4.7. Non-Denaturing Polyacrylamide Gel Electrophoresis
4.8. Fluorescence Assays—miRNA Detection
4.9. Statistic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinstein, I.B.; Case, K. The History of Cancer Research: Introducing an AACR Centennial Series. Cancer Res. 2008, 68, 6861–6862. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bade, B.C.; Cruz, C.S.D. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin. Chest Med. 2020, 41, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [Green Version]
- Sekido, Y.; Fong, K.M.; Minna, J.D. Molecular Genetics of Lung Cancer. Annu. Rev. Med. 2003, 54, 73–87. [Google Scholar] [CrossRef]
- Tsim, S.; O’Dowd, C.; Milroy, R.; Davidson, S. Staging of non-small cell lung cancer (NSCLC): A review. Respir. Med. 2010, 104, 1767–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duruisseaux, M.; Esteller, M. Lung cancer epigenetics: From knowledge to applications. Semin. Cancer Biol. 2018, 51, 116–128. [Google Scholar] [CrossRef]
- Zheng, M. Classification and Pathology of Lung Cancer. Surg. Oncol. Clin. N. Am. 2016, 25, 447–468. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Athey, V.L.; Suckling, R.J.; Tod, A.; Walters, S.; Rogers, T.K. Early diagnosis of lung cancer: Evaluation of a community-based social marketing intervention. Thorax 2011, 67, 412–417. [Google Scholar] [CrossRef] [Green Version]
- Knight, S.B.; Phil, A.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dent, A.G.; Sutedja, T.G.; Zimmerman, P.V. Exhaled breath analysis for lung cancer. J. Thorac. Dis. 2013, 5, S540–S550. [Google Scholar] [CrossRef] [PubMed]
- Villalobos, P.; Wistuba, I.I. Lung Cancer Biomarkers. Hematol. Clin. N. Am. 2017, 31, 13–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, J.; Shen, J.; Leng, Q.; Qin, M.; Zhan, M.; Jiang, F. MicroRNA-based biomarkers for diagnosis of non-small cell lung cancer (NSCLC). Thorac. Cancer 2020, 11, 762–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Li, H. miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC). J. Thorac. Dis. 2018, 10, 3119–3131. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achieve Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.-L.; Tsai, Y.-M.; Lien, C.-T.; Kuo, P.-L.; Hung, A.J.-Y. The Roles of MicroRNA in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Kudo, M.; Huang, X.; Sui, H.; Tian, H.; Croce, C.M.; Cui, R. Frontiers of MicroRNA Signature in Non-small Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 643942. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Meng, F.; Chan, L.W.C.; Cho, W.C.S.; Wong, S.C.C. Circulating Plasma MicroRNAs As Diagnostic Markers for NSCLC. Front. Genet. 2016, 7, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanafi, A.R.; Jayusman, A.M.; Alfasunu, S.; Sadewa, A.H.; Pramono, D.; Heriyanto, D.S.; Haryana, S.M. Serum MiRNA as Predictive and Prognosis Biomarker in Advanced Stage Non-small Cell Lung Cancer in Indonesia. Zhongguo Fei Ai Za Zhi = Chin. J. Lung Cancer 2020, 23, 321–332. [Google Scholar] [CrossRef]
- Salem, M.L.; Okba, S.; Morad, S.; El-Desoky, N.; Barakat, A.F. Altered expression of miRNA-155 and miRNA-21 and higher numbers of T cells in the peripheral blood of early diagnosed lung cancer patients. Int. J. Cancer Biomed. Res. 2020, 4, 19–25. [Google Scholar] [CrossRef]
- Mosallaei, M.; Ehtesham, N.; Rahimirad, S.; Saghi, M.; Vatandoost, N.; Khosravi, S. PBMCs: A new source of diagnostic and prognostic biomarkers. Arch. Physiol. Biochem. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Wan-Ling, H.; Hsieh, W.-L.; Lai, H.-W.; Tsai, C.-H.; Cheng, Y.-W. MicroRNA Expression Profiling in PBMCs: A Potential Diagnostic Biomarker of Chronic Hepatitis C. Dis. Markers 2014, 2014, 367157. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, S.; Sun, G.; Tang, X.; Lu, S.; Neyrolles, O.; Gao, Q. Comparative miRNA Expression Profiles in Individuals with Latent and Active Tuberculosis. PLoS ONE 2011, 6, e25832. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wu, L.; Mo, X.; Xia, W.; Guo, Y.; Wang, M.; Zeng, K.; Wu, J.; Qiu, Y.; Lin, X.; et al. Identification of PBMC-expressed miRNAs for rheumatoid arthritis. Epigenetics 2020, 15, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lin, Y.; Zhan, M.; Mann, D.L.; Stass, S.A.; Jiang, F. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab. Investig. 2015, 95, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Wang, K.; Drake, T.J. Molecular beacons. Curr. Opin. Chem. Biol. 2004, 8, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Tang, Z.; Yang, C.J.; Kim, Y.; Fang, X.; Li, W.; Wu, Y.; Medley, C.D.; Cao, Z.; Li, J.; et al. Molecular Engineering of DNA: Molecular Beacons. Angew. Chem. Int. Ed. 2009, 48, 856–870. [Google Scholar] [CrossRef] [Green Version]
- Miranda, A.; Santos, T.; Carvalho, J.; Alexandre, D.; Jardim, A.; Caneira, C.; Vaz, V.; Pereira, B.; Godinho, R.; Brito, D.; et al. Aptamer-based approaches to detect nucleolin in prostate cancer. Talanta 2021, 226, 122037. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Lopes-Nunes, J.; Figueiredo, J.; Santos, T.; Miranda, A.; Riscado, M.; Sousa, F.; Duarte, A.P.; Socorro, S.; Tomaz, C.T.; et al. Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection. Sensors 2021, 21, 7015. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, J.A.; Kwon, M.H.; Kang, J.Y.; Rhee, W.J. In situ single step detection of exosome microRNA using molecular beacon. Biomaterials 2015, 54, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Zhang, A.-M.; Ma, H.; Lin, S.; Wang, X.-X.; Sun, J.-G.; Chen, Z.-T. Novel molecular beacons to monitor microRNAs in non-small-cell lung cancer. Mol. Cell. Probes 2012, 26, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fan, J.; Zhou, C.; Wang, L.; Zhao, B.; Zhang, H.; Liu, B.; Tong, C. Quantitative Detection of miRNA-21 Expression in Tumor Cells and Tissues Based on Molecular Beacon. Int. J. Anal. Chem. 2018, 2018, 3625823. [Google Scholar] [CrossRef] [Green Version]
- Shahsavar, K.; Shokri, E.; Hosseini, M. A fluorescence-readout method for miRNA-155 detection with double-hairpin molecular beacon based on quadruplex DNA structure. Microchem. J. 2020, 158, 105277. [Google Scholar] [CrossRef]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life Sci. 2018, 75, 3539–3551. [Google Scholar] [CrossRef]
- Zhang, J.-G.; Wang, J.-J.; Zhao, F.; Liu, Q.; Jiang, K.; Yang, G.-H. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 2010, 411, 846–852. [Google Scholar] [CrossRef]
- Liu, Z.-L.; Wang, H.; Liu, J.; Wang, Z.-X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell. Biochem. 2012, 372, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhu, F.; Liu, J.; Xu, T.; Pei, D.; Wang, R.; Qian, Y.; Li, Q.; Wang, L.; Shi, Z.; et al. Alteration in Mir-21/PTEN Expression Modulates Gefitinib Resistance in Non-Small Cell Lung Cancer. PLoS ONE 2014, 9, e103305. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, D.; Sun, G.; Mei, F.; Cui, Y.; Xu, H. Effect of miR-21 on Apoptosis in Lung Cancer Cell Through Inhibiting the PI3K/ Akt/NF-κB Signaling Pathway in Vitro and in Vivo. Cell. Physiol. Biochem. 2018, 46, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Mohamadnia, A.; Azimi, S.A.; Nia, H.R.S.; Jamaati, H.R.; Dargahi, H.; Pasdar, A.; Khosravi, A.; Dizaji, M.K.; Bahrami, N.; Daghighi, S.; et al. Ectopic Expression of miRNA-21 and miRNA-205 in Non-Small Cell Lung Cancer. Int. J. Cancer Manag. 2019, 12, e85456. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Guan, Y.; Liu, X.; Meng, Q.; Guo, Q. MiR-92b regulates the cell growth, cisplatin chemosensitivity of A549 non small cell lung cancer cell line and target PTEN. Biochem. Biophys. Res. Commun. 2013, 440, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-T.; Ma, Z.-L.; Li, Y.-L.; Wang, Y.-Q.; Zhao, B.-T.; Wei, J.-L.; Qiu, X.; Zhao, X.-T.; Jin, Y.-X.; Qi, X. miR-150, p53 protein and relevant miRNAs consist of a regulatory network in NSCLC tumorigenesis. Oncol. Rep. 2013, 30, 492–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, X.; Liu, Y.; Wang, Y.; Meng, M.; Wang, K.; Zang, X.; Zhao, S.; Sun, X.; Cui, L.; Pan, L.; et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 2016, 7, 84508–84519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braicu, C.; Gulei, D.; Cojocneanu, R.; Raduly, L.; Jurj, A.; Knutsen, E.; Calin, G.A.; Berindan-Neagoe, I. miR-181a/b therapy in lung cancer: Reality or myth? Mol. Oncol. 2019, 13, 9–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powrózek, T.; Krawczyk, P.; Kowalski, D.; Winiarczyk, K.; Olszyna-Serementa, M.; Milanowski, J. Plasma circulating microRNA-944 and microRNA-3662 as potential histologic type-specific early lung cancer biomarkers. Transl. Res. 2015, 166, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Ma, J.; Wang, Y.; Zan, J.; Wang, Z.; Zhu, Y.; Zhu, Y.; Ling, L.; Cao, L.; Liu, X.; et al. miR-21-5p induces cell proliferation by targeting TGFBI in non-small cell lung cancer cells. Exp. Ther. Med. 2018, 16, 4655–4663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farahani, N.; Behmanesh, M.; Ranjbar, B. Evaluation of Rationally Designed Label-free Stem-loop DNA Probe Opening in the Presence of miR-21 by Circular Dichroism and Fluorescence Techniques. Sci. Rep. 2020, 10, 4018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikuta, S.; Chattopadhyaya, R.; Ito, H.; Dickerson, R.E.; Kearns, D.R. NMR study of a synthetic DNA hairpin. Biochemistry 1986, 25, 4840–4849. [Google Scholar] [CrossRef]
- Ghosh, M.; Chary, K.V.R.; Kumar, N.V.; Varshney, U. Structural characterisation of a uracil containing hairpin DNA by NMR and molecular dynamics. Nucleic Acids Res. 1999, 27, 3938–3944. [Google Scholar] [CrossRef] [Green Version]
- Davis, P.W.; Thurmes, W.; Tinoco, L. Structure of a small RNA hairpin. Nucleic Acids Res. 1993, 21, 537–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirayil, S.; Wu, Q.; Amezcua, C.; Luebke, K.J. NMR Characterization of an Oligonucleotide Model of the MiR-21 Pre-Element. PLoS ONE 2014, 9, e108231. [Google Scholar] [CrossRef]
- Ramesh, V. NMR evidence for the RNA stem-loop structure involved in the transcription attenuation of E.coli trp operon. Nucleic Acids Res. 1993, 21, 5485–5488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]




| NSCLC (N = 50) | HEALTHY INDIVIDUALS (N = 22) | |
|---|---|---|
| AGE (YEARS) | ||
| Mean ± SD | 69.92 ± 8.08 | 29.5 ± 8.07 |
| GENDER | ||
| Male | 37 (74%) | 9 (41%) |
| Female | 13 (26%) | 13 (59%) |
| SMOKING STATUS | ||
| Nonsmoker | 20 (40%) | 19 (86%) |
| Smoker | 16 (32%) | 3 (14%) |
| Ex-smoker | 14 (28%) | 0 (0%) |
| HISTOLOGICAL TYPE | ||
| Adenocarcinoma | 36 (72%) | NA |
| Squamous cell carcinoma | 10 (20%) | NA |
| Large cell carcinoma | 4 (8%) | NA |
| TNM STAGE | ||
| I | 2 (4%) | NA |
| II | 4 (8%) | NA |
| III | 8 (16%) | NA |
| IV | 36 (72%) | NA |
| METASTASIS | ||
| Yes | 33 (66%) | NA |
| No | 17 (34%) | NA |
| Name (miRCURY Primers) | Role | miRNA Target Sequence (from 5′ to 3′) |
|---|---|---|
| hsa-miR-103a-3p | Housekeeping | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-191-5p | Housekeeping | CAACGGAAUCCCAAAAGCAGCUG |
| U6snRNA hsa-miR-21-3p | Housekeeping NSCLC-related | CACGAATTTGCGTGTCATCCTT CAACACCAGUCGAUGGGCUGU |
| hsa-miR-21-5p | NSCLC-related | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-92b-5p | NSCLC-related | AGGGACGGGACGCGGUGCAGUG |
| hsa-miR-150-3p | NSCLC-related | CUGGUACAGGCCUGGGGGACAG |
| hsa-miR-155-3p | NSCLC-related | CUCCUACAUAUUAGCAUUAACA |
| hsa-miR-155-5p | NSCLC-related | UUAAUGCUAAUCGUGAUAGGGGUU |
| hsa-miR-181a-5p | NSCLC-related | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-3662 | NSCLC-related | GAAAAUGAUGAGUAGUGACUGAUG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandre, D.; Teixeira, B.; Rico, A.; Valente, S.; Craveiro, A.; Baptista, P.V.; Cruz, C. Molecular Beacon for Detection miRNA-21 as a Biomarker of Lung Cancer. Int. J. Mol. Sci. 2022, 23, 3330. https://doi.org/10.3390/ijms23063330
Alexandre D, Teixeira B, Rico A, Valente S, Craveiro A, Baptista PV, Cruz C. Molecular Beacon for Detection miRNA-21 as a Biomarker of Lung Cancer. International Journal of Molecular Sciences. 2022; 23(6):3330. https://doi.org/10.3390/ijms23063330
Chicago/Turabian StyleAlexandre, Daniela, Bernardo Teixeira, André Rico, Salete Valente, Ana Craveiro, Pedro V. Baptista, and Carla Cruz. 2022. "Molecular Beacon for Detection miRNA-21 as a Biomarker of Lung Cancer" International Journal of Molecular Sciences 23, no. 6: 3330. https://doi.org/10.3390/ijms23063330
APA StyleAlexandre, D., Teixeira, B., Rico, A., Valente, S., Craveiro, A., Baptista, P. V., & Cruz, C. (2022). Molecular Beacon for Detection miRNA-21 as a Biomarker of Lung Cancer. International Journal of Molecular Sciences, 23(6), 3330. https://doi.org/10.3390/ijms23063330