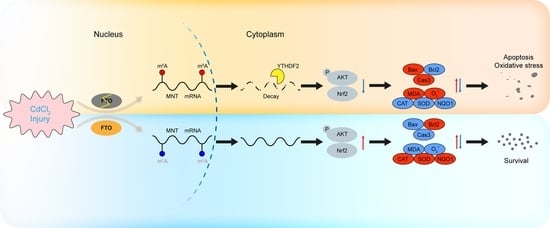
FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells
Abstract
:1. Introduction
2. Results
2.1. Cd Increased the Degree of Apoptosis in Granulosa Cells
2.2. Cd-Induced Oxidative Stress in Granulosa Cells
2.3. Cd Alters the Expression of m6A Methylation-Regulated Genes in Granulosa Cells
2.4. Transcriptome and MeRIP Sequencing to Identified m6A modification Targets
2.5. FTO Affects m6A Modification and Expression Level of the MNT mRNA
2.6. FTO Overexpression Suppressed Cd-Induced Apoptosis, Perhaps by Activating the AKT Pathway in Granulosa Cells
2.7. FTO Overexpression Suppressed Cd-Induced Oxidative Stress
2.8. siMNT Reverses the Protective Effect of FTO during Cd Injury in Granulosa Cells
3. Discussion
4. Materials and Methods
4.1. Primary Cell Culture
4.2. Cell Viability
4.3. Transient Cell Transfection
4.4. Real-Time Quantitative PCR (RT-qPCR)
4.5. Western Blot Analysis
4.6. Apoptosis Assay
4.7. TUNEL Assay
4.8. ELISA
4.9. ROS Assay
4.10. Transcriptome Sequencing and MeRIP Sequencing Assay
4.11. Bioinformatics
4.12. MeRIP-RT-qPCR
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jaffe, L.A.; Egbert, J.R. Regulation of Mammalian Oocyte Meiosis by Intercellular Communication within the Ovarian Follicle. Annu. Rev. Physiol. 2017, 79, 237–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.F.; Mokhtar, M.B. Assessing Cadmium and Chromium Concentrations in Drinking Water to Predict Health Risk in Malaysia. Int. J. Environ. Res. Public Health 2020, 17, 2966. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Teng, Y.; Lu, S.; Wang, Y.; Wang, J. Contamination features and health risk of soil heavy metals in China. Sci. Total Environ. 2015, 512, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, N.T.M.; Chuyen, N.V.; Thao, N.T.T.; Duc, N.Q.; Trang, N.T.T.; Binh, N.T.T.; Sa, H.C.; Tran, N.B.; Ba, N.V.; Khai, N.V.; et al. Chromium, Cadmium, Lead, and Arsenic Concentrations in Water, Vegetables, and Seafood Consumed in a Coastal Area in Northern Vietnam. Environ. Health Insights 2020, 14, 1178630220921410. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, N.; Su, C.; Wang, J.; Soyeurt, H. Relationships between Pb, As, Cr, and Cd in individual cows’ milk and milk composition and heavy metal contents in water, silage, and soil. Environ. Pollut. 2019, 255, 113322. [Google Scholar] [CrossRef]
- Hashemi, M. Heavy metal concentrations in bovine tissues (muscle, liver and kidney) and their relationship with heavy metal contents in consumed feed. Ecotoxicol. Environ. Saf. 2018, 154, 263–267. [Google Scholar] [CrossRef]
- Suprewicz, K.; Kozikowska, I.; Chrobaczyńska-Dylag, M.; Gał, A.; Piekarz, A.; Sikora, J.; Sławska, H.; Stawarz, R. Effects of the cigarette smoking on the newborn clinical parametrs and the accumulation of cadmium and lead in the placenta of women from Upper Silesia. Ginekol. Pol. 2013, 84, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Richter, P.A.; Bishop, E.E.; Wang, J.; Swahn, M.H. Tobacco smoke exposure and levels of urinary metals in the U.S. youth and adult population: The National Health and Nutrition Examination Survey (NHANES) 1999–2004. Int. J. Environ. Res. Public Health 2009, 6, 1930–1946. [Google Scholar] [CrossRef]
- Liu, J.; Luo, L.F.; Wang, D.L.; Wang, W.X.; Zhu, J.L.; Li, Y.C.; Chen, N.Z.; Huang, H.L.; Zhang, W.C. Cadmium induces ovarian granulosa cell damage by activating PERK-eIF2α-ATF4 through endoplasmic reticulum stress. Biol. Reprod. 2019, 100, 292–299. [Google Scholar] [CrossRef]
- Wan, X.; Zhu, J.; Zhu, Y.; Zhu, Y.; Ma, X.; Zheng, Y.; Wang, F.; Liu, Z.; Zhang, T. Rat ovarian follicle bioassay reveals adverse effects of cadmium chloride (CdCl2) exposure on follicle development and oocyte maturation. Toxicol. Ind. Health 2010, 26, 609–618. [Google Scholar] [CrossRef]
- Zhang, W.; Jia, H. Effect and mechanism of cadmium on the progesterone synthesis of ovaries. Toxicology 2007, 239, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.; Wang, W.; Li, Y.; Li, H.; Lu, X.; Xiao, S.; Wu, T.; Xie, M.; Zhang, W. Continuous cadmium exposure from weaning to maturity induces downregulation of ovarian follicle development-related SCF/c-kit gene expression and the corresponding changes of DNA methylation/microRNA pattern. Toxicol. Lett. 2014, 225, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ma, W.; Su, Y. NF-κB pathway contributes to cadmium-induced apoptosis of porcine granulosa cells. Biol. Trace Elem. Res. 2013, 153, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Liaño-Pons, J.; Arsenian-Henriksson, M.; León, J. The Multiple Faces of MNT and Its Role as a MYC Modulator. Cancers 2021, 13, 4682. [Google Scholar] [CrossRef]
- Toyo-oka, K.; Hirotsune, S.; Gambello, M.J.; Zhou, Z.Q.; Olson, L.; Rosenfeld, M.G.; Eisenman, R.; Hurlin, P.; Wynshaw-Boris, A. Loss of the Max-interacting protein Mnt in mice results in decreased viability, defective embryonic growth and craniofacial defects: Relevance to Miller-Dieker syndrome. Hum. Mol. Genet. 2004, 13, 1057–1067. [Google Scholar] [CrossRef] [Green Version]
- Hurlin, P.J.; Zhou, Z.Q.; Toyo-oka, K.; Ota, S.; Walker, W.L.; Hirotsune, S.; Wynshaw-Boris, A. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J. 2003, 22, 4584–4596. [Google Scholar] [CrossRef] [Green Version]
- Hurlin, P.J.; Quéva, C.; Eisenman, R.N. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997, 11, 44–58. [Google Scholar] [CrossRef] [Green Version]
- Meroni, G.; Reymond, A.; Alcalay, M.; Borsani, G.; Tanigami, A.; Tonlorenzi, R.; Lo Nigro, C.; Messali, S.; Zollo, M.; Ledbetter, D.H.; et al. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997, 16, 2892–2906. [Google Scholar] [CrossRef]
- Eischen, C.M.; Woo, D.; Roussel, M.F.; Cleveland, J.L. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol. Cell. Biol. 2001, 21, 5063–5070. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottman, F.M.; Desrosiers, R.C.; Friderici, K. Nucleotide methylation patterns in eukaryotic mRNA. Prog. Nucleic Acid Res. Mol. Biol. 1976, 19, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Schibler, U.; Kelley, D.E.; Perry, R.P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977, 115, 695–714. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6A(m), and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5. [Google Scholar] [CrossRef] [Green Version]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Shen, Z.; Liu, P.; Sun, Q.; Li, Y.; Acharya, R.; Li, X.; Sun, C. FTO inhibits UPR(mt)-induced apoptosis by activating JAK2/STAT3 pathway and reducing m6A level in adipocytes. Apoptosis Int. J. Program. Cell Death 2021, 26, 474–487. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, J.; Wu, Y.; Han, L.; Chen, J.; Wei, Y.; Shen, L.; Long, C.; Wu, S.; Wei, G. Increased m6A modification of RNA methylation related to the inhibition of demethylase FTO contributes to MEHP-induced Leydig cell injury(☆). Environ. Pollut. 2021, 268, 115627. [Google Scholar] [CrossRef]
- Tang, J.; Su, Q.; Guo, Z.; Zhou, J.; Zheng, F.; Yu, G.; Shao, W.; Hu, H.; Wu, S.; Li, H. N6-methyladenosine(m6A) demethylase FTO regulates cellular apoptosis following cobalt-induced oxidative stress. Environ. Pollut. 2022, 297, 118749. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Han, X.; Li, W.; Wang, N.; Yao, L.; Zhao, Y.; Zhang, L. N6-methyladenosine Demethylase FTO Induces the Dysfunctions of Ovarian Granulosa Cells by Upregulating Flotillin 2. Reprod. Sci. 2022, 29, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.X.; Wang, Y.N.; Li, Z.Y.; Dai, Z.H.; He, Y.; Chu, K.; Gu, J.Y.; Ji, Y.X.; Sun, N.X.; Yang, F.; et al. The m6A mRNA demethylase FTO in granulosa cells retards FOS-dependent ovarian aging. Cell Death Dis. 2021, 12, 744. [Google Scholar] [CrossRef]
- Zhao, L.; Kong, X.; Zhong, W.; Wang, Y.; Li, P. FTO accelerates ovarian cancer cell growth by promoting proliferation, inhibiting apoptosis, and activating autophagy. Pathol. Res. Pract. 2020, 216, 153042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.X.; Wang, J.K.; Shen, L.J.; Long, C.L.; Liu, B.; Wei, Y.; Han, L.D.; Wei, Y.X.; Wu, S.D.; Wei, G.H. Increased m6A RNA modification is related to the inhibition of the Nrf2-mediated antioxidant response in di-(2-ethylhexyl) phthalate-induced prepubertal testicular injury. Environ. Pollut. 2020, 259, 113911. [Google Scholar] [CrossRef]
- Liu, S.; Zhuo, L.; Wang, J.; Zhang, Q.; Li, Q.; Li, G.; Yan, L.; Jin, T.; Pan, T.; Sui, X.; et al. METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 2020, 10, 1631–1646. [Google Scholar] [PubMed]
- Xu, G.; Liu, S.; Huang, M.; Jiang, X.; Yang, M. Cadmium induces apoptosis of human granulosa cell line KGN via mitochondrial dysfunction-mediated pathways. Ecotoxicol. Environ. Saf. 2021, 220, 112341. [Google Scholar] [CrossRef]
- Zhu, M.; Miao, S.; Zhou, W.; Elnesr, S.S.; Dong, X.; Zou, X. MAPK, AKT/FoxO3a and mTOR pathways are involved in cadmium regulating the cell cycle, proliferation and apoptosis of chicken follicular granulosa cells. Ecotoxicol. Environ. Saf. 2021, 214, 112091. [Google Scholar] [CrossRef]
- Ren, X.; Wang, S.; Zhang, C.; Hu, X.; Zhou, L.; Li, Y.; Xu, L. Selenium ameliorates cadmium-induced mouse leydig TM3 cell apoptosis via inhibiting the ROS/JNK /c-jun signaling pathway. Ecotoxicol. Environ. Saf. 2020, 192, 110266. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Yang, L.; Xu, H.; Wu, Y.; Wu, J.; Chen, L.; Xu, C. Magnesium isoglycyrrhizinate prevents cadmium-induced activation of JNK and apoptotic hepatocyte death by reversing ROS-inactivated PP2A. J. Pharm. Pharmacol. 2021, 73, 1663–1674. [Google Scholar] [CrossRef]
- Yiming, Z.; Zhaoyi, L.; Jing, L.; Jinliang, W.; Zhiqiang, S.; Guangliang, S.; Shu, L. Cadmium induces the thymus apoptosis of pigs through ROS-dependent PTEN/PI3K/AKT signaling pathway. Environ. Sci. Pollut. Res. Int. 2021, 28, 39982–39992. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zong, C.; Liu, J.; Zeng, L.; Li, Q.; Liu, Z.; Li, Y.; Zhu, J.; Li, L.; Zhang, C.; et al. C-myc promotes miR-92a-2-5p transcription in rat ovarian granulosa cells after cadmium exposure. Toxicol. Appl. Pharmacol. 2021, 421, 115536. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, M.; Chen, B.; Wang, Q.; Pan, S.; Hou, Y.; Xia, J.; Zhou, X. ALKBH5 promotes cadmium-induced transformation of human bronchial epithelial cells by regulating PTEN expression in an m6A-dependent manner. Ecotoxicol. Environ. Saf. 2021, 224, 112686. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, T.W.; Ko, K.S.; Xia, M.; Lu, S.C. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology 2009, 49, 860–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Link, J.M.; Ota, S.; Zhou, Z.Q.; Daniel, C.J.; Sears, R.C.; Hurlin, P.J. A critical role for Mnt in Myc-driven T-cell proliferation and oncogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 19685–19690. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Bashir, N.; Shagirtha, K.; Manoharan, V.; Miltonprabu, S. The molecular and biochemical insight view of grape seed proanthocyanidins in ameliorating cadmium-induced testes-toxicity in rat model: Implication of PI3K/Akt/Nrf-2 signaling. Biosci. Rep. 2019, 39, BSR20180515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terragni, J.; Nayak, G.; Banerjee, S.; Medrano, J.L.; Graham, J.R.; Brennan, J.F.; Sepulveda, S.; Cooper, G.M. The E-box binding factors Max/Mnt, MITF, and USF1 act coordinately with FoxO to regulate expression of proapoptotic and cell cycle control genes by phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3 signaling. J. Biol. Chem. 2011, 286, 36215–36227. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Luo, S.; Fan, P.; Zhu, H.; Li, Y.; Huang, W. Growth hormone activates PI3K/Akt signaling and inhibits ROS accumulation and apoptosis in granulosa cells of patients with polycystic ovary syndrome. Reprod. Biol. Endocrinol. RBE 2020, 18, 121. [Google Scholar] [CrossRef]
- Wang, S.; Lin, S.; Zhu, M.; Li, C.; Chen, S.; Pu, L.; Lin, J.; Cao, L.; Zhang, Y. Acupuncture Reduces Apoptosis of Granulosa Cells in Rats with Premature Ovarian Failure Via Restoring the PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 6311. [Google Scholar] [CrossRef] [Green Version]
- Akino, N.; Wada-Hiraike, O.; Terao, H.; Honjoh, H.; Isono, W.; Fu, H.; Hirano, M.; Miyamoto, Y.; Tanikawa, M.; Harada, M.; et al. Activation of Nrf2 might reduce oxidative stress in human granulosa cells. Mol. Cell. Endocrinol. 2018, 470, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Khadrawy, O.; Gebremedhn, S.; Salilew-Wondim, D.; Taqi, M.O.; Neuhoff, C.; Tholen, E.; Hoelker, M.; Schellander, K.; Tesfaye, D. Endogenous and Exogenous Modulation of Nrf2 Mediated Oxidative Stress Response in Bovine Granulosa Cells: Potential Implication for Ovarian Function. Int. J. Mol. Sci. 2019, 20, 1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.; Zhao, J.; Liu, H.; Wang, J.; Lu, W. BMAL1 knockdown promoted apoptosis and reduced testosterone secretion in TM3 Leydig cell line. Gene 2020, 747, 144672. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Zhang, J.B.; Zheng, Y.; Zhang, W.D.; Guo, H.X.; Cong, S.; Ding, Y.; Yuan, B. Comprehensive analysis of differences in N6-methyladenosine RNA methylomes in the rat adenohypophysis after GnRH treatment. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22204. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Lu, Z.; Liu, H.; Zhang, L.; Zhang, S.; Chen, Y.; Rao, M.K.; Huang, Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods 2014, 69, 274–281. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, H.; Li, Z.; Li, X.; Yang, X.; Zhao, J.; Guo, J.; Lu, W.; Liu, H.; Wang, J. FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells. Int. J. Mol. Sci. 2022, 23, 4948. https://doi.org/10.3390/ijms23094948
Ding H, Li Z, Li X, Yang X, Zhao J, Guo J, Lu W, Liu H, Wang J. FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells. International Journal of Molecular Sciences. 2022; 23(9):4948. https://doi.org/10.3390/ijms23094948
Chicago/Turabian StyleDing, He, Zhiqiang Li, Xin Li, Xiaorui Yang, Jing Zhao, Jing Guo, Wenfa Lu, Hongyu Liu, and Jun Wang. 2022. "FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells" International Journal of Molecular Sciences 23, no. 9: 4948. https://doi.org/10.3390/ijms23094948
APA StyleDing, H., Li, Z., Li, X., Yang, X., Zhao, J., Guo, J., Lu, W., Liu, H., & Wang, J. (2022). FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells. International Journal of Molecular Sciences, 23(9), 4948. https://doi.org/10.3390/ijms23094948