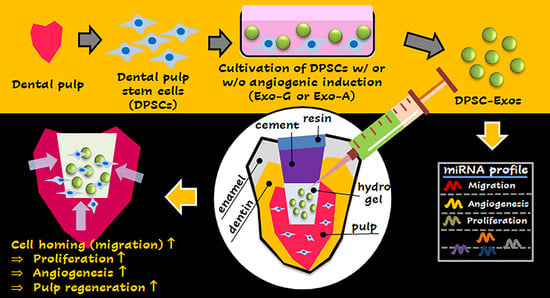
Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration
Abstract
:1. Introduction
2. Results
2.1. Characterization of DPSCs and DPSC-Exos
2.2. Effect of DPSC-Exos on Cell Toxicity, Proliferation, Migration, and Angiogenic Differentiation
2.3. Exosomal miRNA Profile
3. Discussion
4. Materials and Methods
4.1. Isolation of DPSCs
4.2. Characterization of DPSCs
4.3. Isolation of DPSC-Exos
4.4. Nanoparticle Tracking Analyzer (NTA)
4.5. Exosome Antibody Array
4.6. Scanning Electron Microscopy (SEM)
4.7. Exosome Labeling and Cellular Uptake
4.8. Cell Toxicity
4.9. Cell Proliferation
4.10. Cell Migration
4.11. Angiogenic Differentiation
4.12. Real-Time Polymerase Chain Reaction (RT-PCR)
4.13. Next-Generation Sequencing (NGS)
4.14. Statistics
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Islam, B.; Khan, S.N.; Khan, A.U. Dental caries: From infection to prevention. Med. Sci. Monit. 2007, 13, RA196–RA203. [Google Scholar] [PubMed]
- Foreman, P.C.; Barnes, I.E. Review of calcium hydroxide. Int. Endod. J. 1990, 23, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Mass, E.; Zilberman, U. Long-term radiologic pulp evaluation after partial pulpotomy in young permanent molars. Quintessence Int. 2011, 42, 547–554. [Google Scholar] [PubMed]
- Parirokh, M.; Torabinejad, M. Mineral trioxide aggregate: A comprehensive literature review—Part I: Chemical, physical, and antibacterial properties. J. Endod. 2010, 36, 16–27. [Google Scholar] [CrossRef]
- Torabinejad, M.; Parirokh, M. Mineral trioxide aggregate: A comprehensive literature review—Part II: Leakage and biocompatibility investigations. J. Endod. 2010, 36, 190–202. [Google Scholar] [CrossRef]
- Desai, S.; Chandler, N. Calcium hydroxide-based root canal sealers: A review. J. Endod. 2009, 35, 475–480. [Google Scholar] [CrossRef]
- Parirokh, M.; Torabinejad, M. Mineral trioxide aggregate: A comprehensive literature review—Part III: Clinical applications, drawbacks, and mechanism of action. J. Endod. 2010, 36, 400–413. [Google Scholar] [CrossRef]
- Ghoddusi, J.; Forghani, M.; Parisay, I. New approaches in vital pulp therapy in permanent teeth. Iran Endod. J. 2014, 9, 15–22. [Google Scholar]
- Ward, J. Vital pulp therapy in cariously exposed permanent teeth and its limitations. Aust. Endod. J. 2002, 28, 29–37. [Google Scholar] [CrossRef]
- Iohara, K.; Imabayashi, K.; Ishizaka, R.; Watanabe, A.; Nabekura, J.; Ito, M.; Matsushita, K.; Nakamura, H.; Nakashima, M. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue Eng. Part A 2011, 17, 1911–1920. [Google Scholar] [CrossRef]
- Ishizaka, R.; Iohara, K.; Murakami, M.; Fukuta, O.; Nakashima, M. Regeneration of dental pulp following pulpectomy by fractionated stem/progenitor cells from bone marrow and adipose tissue. Biomaterials 2012, 33, 2109–2118. [Google Scholar] [CrossRef] [PubMed]
- Xuan, K.; Li, B.; Guo, H.; Sun, W.; Kou, X.; He, X.; Zhang, Y.; Sun, J.; Liu, A.; Liao, L.; et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci. Transl. Med. 2018, 10, eaaf3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, J.J.; Kim, S.G.; Zhou, J.; Ye, L.; Cho, S.; Suzuki, T.; Fu, S.Y.; Yang, R.; Zhou, X. Regenerative endodontics: Barriers and strategies for clinical translation. Dent. Clin. N. Am. 2012, 56, 639–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eramo, S.; Natali, A.; Pinna, R.; Milia, E. Dental pulp regeneration via cell homing. Int. Endod. J. 2018, 51, 405–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.T.; Gronthos, S.; Shi, S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: Their biology and role in regenerative medicine. J. Dent. Res. 2009, 88, 792–806. [Google Scholar] [CrossRef]
- Shi, X.; Mao, J.; Liu, Y. Concise review: Pulp stem cells derived from human permanent and deciduous teeth: Biological characteristics and therapeutic applications. Stem Cells Transl. Med. 2020, 9, 445–464. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, M.; Farges, J.C.; Lacerda-Pinheiro, S.; Six, N.; Jegat, N.; Decup, F.; Septier, D.; Carrouel, F.; Durand, S.; Chaussain-Miller, C.; et al. Inflammatory and immunological aspects of dental pulp repair. Pharmacol. Res. 2008, 58, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.C.; Yuan, T.; Zhang, Y.L.; Yin, W.J.; Guo, S.C.; Zhang, C.Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef]
- Tetta, C.; Consiglio, A.L.; Bruno, S.; Tetta, E.; Gatti, E.; Dobreva, M.; Cremonesi, F.; Camussi, G. The role of microvesicles derived from mesenchymal stem cells in tissue regeneration; a dream for tendon repair? Muscles Ligaments Tendons J. 2012, 2, 212–221. [Google Scholar]
- Zhang, B.; Wang, M.; Gong, A.; Zhang, X.; Wu, X.; Zhu, Y.; Shi, H.; Wu, L.; Zhu, W.; Qian, H.; et al. HucMSC-Exosome Mediated-Wnt4 Signaling Is Required for Cutaneous Wound Healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chu, W.C.; Lai, R.C.; Lim, S.K.; Hui, J.H.; Toh, W.S. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthr. Cartil. 2016, 24, 2135–2140. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Zhang, H. MicroRNAs in the Migration of Mesenchymal Stem Cells. Stem Cell Rev. Rep. 2019, 15, 3–12. [Google Scholar] [CrossRef]
- Tao, S.C.; Guo, S.C.; Zhang, C.Q. Platelet-derived Extracellular Vesicles: An Emerging Therapeutic Approach. Int. J. Biol. Sci. 2017, 13, 828–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.C.; Tao, S.C.; Yin, W.J.; Qi, X.; Yuan, T.; Zhang, C.Q. Exosomes derived from platelet-rich plasma promote the re-epithelization of chronic cutaneous wounds via activation of YAP in a diabetic rat model. Theranostics 2017, 7, 81–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Chuah, S.J.; Lai, R.C.; Hui, J.H.P.; Lim, S.K.; Toh, W.S. MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials 2018, 156, 16–27. [Google Scholar] [CrossRef]
- Hu, X.; Zhong, Y.; Kong, Y.; Chen, Y.; Feng, J.; Zheng, J. Lineage-specific exosomes promote the odontogenic differentiation of human dental pulp stem cells (DPSCs) through TGFbeta1/smads signaling pathway via transfer of microRNAs. Stem Cell Res. Ther. 2019, 10, 170. [Google Scholar] [CrossRef] [Green Version]
- Ivica, A.; Ghayor, C.; Zehnder, M.; Valdec, S.; Weber, F.E. Pulp-Derived Exosomes in a Fibrin-Based Regenerative Root Filling Material. J. Clin. Med. 2020, 9, 491. [Google Scholar] [CrossRef] [Green Version]
- Stanko, P.; Altanerova, U.; Jakubechova, J.; Repiska, V.; Altaner, C. Dental Mesenchymal Stem/Stromal Cells and Their Exosomes. Stem Cells Int. 2018, 2018, 8973613. [Google Scholar] [CrossRef] [Green Version]
- Coughlan, C.; Bruce, K.D.; Burgy, O.; Boyd, T.D.; Michel, C.R.; Garcia-Perez, J.E.; Adame, V.; Anton, P.; Bettcher, B.M.; Chial, H.J.; et al. Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses. Curr. Protoc. Cell Biol. 2020, 88, e110. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, R.W.; Lai, R.C.; Zhang, B.; Tan, S.S.; Yin, Y.; Teh, B.J.; Lim, S.K. Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Adv. Drug Deliv. Rev. 2013, 65, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Sung, B.H.; Parent, C.A.; Weaver, A.M. Extracellular vesicles: Critical players during cell migration. Dev. Cell 2021, 56, 1861–1874. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [Green Version]
- Kriebel, P.W.; Majumdar, R.; Jenkins, L.M.; Senoo, H.; Wang, W.; Ammu, S.; Chen, S.; Narayan, K.; Iijima, M.; Parent, C.A. Extracellular vesicles direct migration by synthesizing and releasing chemotactic signals. J. Cell Biol. 2018, 217, 2891–2910. [Google Scholar] [CrossRef]
- Sung, B.H.; von Lersner, A.; Guerrero, J.; Krystofiak, E.S.; Inman, D.; Pelletier, R.; Zijlstra, A.; Ponik, S.M.; Weaver, A.M. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 2020, 11, 2092. [Google Scholar] [CrossRef]
- Jimenez, L.; Yu, H.; McKenzie, A.J.; Franklin, J.L.; Patton, J.G.; Liu, Q.; Weaver, A.M. Quantitative Proteomic Analysis of Small and Large Extracellular Vesicles (EVs) Reveals Enrichment of Adhesion Proteins in Small EVs. J. Proteome Res. 2019, 18, 947–959. [Google Scholar] [CrossRef]
- Hakulinen, J.; Sankkila, L.; Sugiyama, N.; Lehti, K.; Keski-Oja, J. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J. Cell Biochem. 2008, 105, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Laghezza Masci, V.; Taddei, A.R.; Gambellini, G.; Giorgi, F.; Fausto, A.M. Microvesicles shed from fibroblasts act as metalloproteinase carriers in a 3-D collagen matrix. J. Circ. Biomark. 2016, 5, 1849454416663660. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, R.; Huang, C.C.; Ravindran, S. Hijacking the Cellular Mail: Exosome Mediated Differentiation of Mesenchymal Stem Cells. Stem Cells Int. 2016, 2016, 3808674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronckaers, A.; Hilkens, P.; Fanton, Y.; Struys, T.; Gervois, P.; Politis, C.; Martens, W.; Lambrichts, I. Angiogenic properties of human dental pulp stem cells. PLoS ONE 2013, 8, e71104. [Google Scholar] [CrossRef] [Green Version]
- Hilkens, P.; Fanton, Y.; Martens, W.; Gervois, P.; Struys, T.; Politis, C.; Lambrichts, I.; Bronckaers, A. Pro-angiogenic impact of dental stem cells in vitro and in vivo. Stem Cell Res. 2014, 12, 778–790. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.L.; Xiao, L.; Du, X.L.; Hong, L.; Li, C.L.; Jiao, J.; Li, W.D.; Li, X.Q. MiR-205 promotes endothelial progenitor cell angiogenesis and deep vein thrombosis recanalization and resolution by targeting PTEN to regulate Akt/autophagy pathway and MMP2 expression. J. Cell Mol. Med. 2019, 23, 8493–8504. [Google Scholar] [CrossRef] [Green Version]
- Brunello, G.; Zanotti, F.; Trentini, M.; Zanolla, I.; Pishavar, E.; Favero, V.; Favero, R.; Favero, L.; Bressan, E.; Bonora, M.; et al. Exosomes Derived from Dental Pulp Stem Cells Show Different Angiogenic and Osteogenic Properties in Relation to the Age of the Donor. Pharmaceutics 2022, 14, 908. [Google Scholar] [CrossRef]
- Xie, K.; Cai, Y.; Yang, P.; Du, F.; Wu, K. Upregulating microRNA-874-3p inhibits CXCL12 expression to promote angiogenesis and suppress inflammatory response in ischemic stroke. Am. J. Physiol. Cell Physiol. 2020, 319, C579–C588. [Google Scholar] [CrossRef]
- Lu, G.D.; Cheng, P.; Liu, T.; Wang, Z. BMSC-Derived Exosomal miR-29a Promotes Angiogenesis and Osteogenesis. Front. Cell Dev. Biol. 2020, 8, 608521. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, L.; Zhu, X.; Xu, J.; Jin, R.; Li, G.; Wu, F. MiR-29a modulates the angiogenic properties of human endothelial cells. Biochem. Biophys. Res. Commun. 2013, 434, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.; Papangeli, I.; Park, Y.; Jeong, H.N.; Choi, J.; Kang, H.; Jo, H.N.; Kim, J.; Chun, H.J. A PPARgamma-dependent miR-424/503-CD40 axis regulates inflammation mediated angiogenesis. Sci. Rep. 2017, 7, 2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Z.; Li, B.; Yang, Z.; Fang, H.; Zhang, G.M.; Feng, Z.H.; Huang, B. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS ONE 2009, 4, e7629. [Google Scholar] [CrossRef] [PubMed]
- Ajay Sharma, L.; Love, R.M.; Ali, M.A.; Sharma, A.; Macari, S.; Avadhani, A.; Dias, G.J. Healing response of rat pulp treated with an injectable keratin hydrogel. J. Appl. Biomater. Funct. Mater. 2017, 15, e244–e250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bader, A.G.; Brown, D.; Winkler, M. The promise of microRNA replacement therapy. Cancer Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Eulalio, A.; Mano, M.; Dal Ferro, M.; Zentilin, L.; Sinagra, G.; Zacchigna, S.; Giacca, M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012, 492, 376–381. [Google Scholar] [CrossRef]
- Joris, V.; Gomez, E.L.; Menchi, L.; Lobysheva, I.; Di Mauro, V.; Esfahani, H.; Condorelli, G.; Balligand, J.L.; Catalucci, D.; Dessy, C. MicroRNA-199a-3p and MicroRNA-199a-5p Take Part to a Redundant Network of Regulation of the NOS (NO Synthase)/NO Pathway in the Endothelium. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2345–2357. [Google Scholar] [CrossRef] [Green Version]
- Urbich, C.; Kuehbacher, A.; Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 2008, 79, 581–588. [Google Scholar] [CrossRef] [Green Version]
- Nicoli, S.; Knyphausen, C.P.; Zhu, L.J.; Lakshmanan, A.; Lawson, N.D. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev. Cell 2012, 22, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Song, W.; Teng, L.; Huang, Y. MiRNA 24-3p-rich exosomes functionalized DEGMA-modified hyaluronic acid hydrogels for corneal epithelial healing. Bioact. Mater. 2022; in press. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, A.; Li, X.; Zhang, S.; Liu, S.; Zhao, H.; Wu, S.; Chen, L.; Ma, C.; Zhao, H. MiR-21-5p regulates extracellular matrix degradation and angiogenesis in TMJOA by targeting Spry1. Arthritis Res. Ther. 2020, 22, 99. [Google Scholar] [CrossRef]
- Xie, J.; Wu, W.; Zheng, L.; Lin, X.; Tai, Y.; Wang, Y.; Wang, L. Roles of MicroRNA-21 in Skin Wound Healing: A Comprehensive Review. Front. Pharmacol. 2022, 13, 828627. [Google Scholar] [CrossRef]
- Deng, S.; Zhao, Q.; Zhen, L.; Zhang, C.; Liu, C.; Wang, G.; Zhang, L.; Bao, L.; Lu, Y.; Meng, L.; et al. Neonatal Heart-Enriched miR-708 Promotes Proliferation and Stress Resistance of Cardiomyocytes in Rodents. Theranostics 2017, 7, 1953–1965. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.G.; Xin, B.C.; Wu, D.; Zhou, L.; Wu, H.B.; Gong, W.; Lv, J. miR-140-5p-mediated regulation of the proliferation and differentiation of human dental pulp stem cells occurs through the lipopolysaccharide/toll-like receptor 4 signaling pathway. Eur. J. Oral Sci. 2017, 125, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Wang, S.; Zhao, R.C. Exosomes from mesenchymal stem/stromal cells: A new therapeutic paradigm. Biomark. Res. 2019, 7, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zayed, M.; Iohara, K.; Watanabe, H.; Ishikawa, M.; Tominaga, M.; Nakashima, M. Characterization of stable hypoxia-preconditioned dental pulp stem cells compared with mobilized dental pulp stem cells for application for pulp regenerative therapy. Stem Cell Res. Ther. 2021, 12, 302. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| miRNA | log2(Fold Change) | p-Value | miRNA | log2(Fold Change) | p-Value |
|---|---|---|---|---|---|
| ocu-miR-708-5p | 5.06 | 0.0000 | ocu-miR-146a-5p | −2.73 | 0.0000 |
| ocu-miR-205-5p | 4.24 | 0.0000 | ocu-miR-503-5p | −2.69 | 0.0000 |
| ocu-miR-708-3p | 4.03 | 0.0000 | ocu-miR-20b-5p | −2.63 | 0.0005 |
| ocu-miR-885-5p | 3.85 | 0.0000 | ocu-miR-18a-3p | −1.97 | 0.0003 |
| ocu-miR-24-2-5p | 3.24 | 0.0000 | ocu-miR-18a-5p | −1.92 | 0.0003 |
| ocu-miR-29c-3p | 3.07 | 0.0000 | ocu-miR-122-5p | −1.91 | 0.0223 |
| ocu-let-7i-3p | 3.04 | 0.0001 | ocu-miR-421-3p | −1.81 | 0.0019 |
| ocu-miR-574-3p | 2.92 | 0.0000 | ocu-miR-350-5p | −1.72 | 0.0099 |
| ocu-miR-874-3p | 2.88 | 0.0000 | ocu-miR-1307-3p | −1.69 | 0.0013 |
| ocu-miR-134-5p | 2.50 | 0.0003 | ocu-miR-98-5p | −1.68 | 0.0004 |
| ocu-miR-125b-5p | 2.20 | 0.0000 | ocu-miR-432-5p | −1.64 | 0.0147 |
| ocu-miR-34c-3p | 2.06 | 0.0006 | ocu-miR-100-5p | −1.60 | 0.0005 |
| ocu-miR-140-3p | 1.99 | 0.0002 | ocu-miR-34a-5p | −1.58 | 0.0094 |
| ocu-miR-342-3p | 1.90 | 0.0000 | ocu-miR-16b-5p | −1.51 | 0.0008 |
| ocu-miR-145-5p | 1.82 | 0.0008 | ocu-miR-20a-5p | −1.50 | 0.0271 |
| ocu-miR-150-5p | 1.63 | 0.0053 | ocu-miR-30a-5p | −1.46 | 0.0004 |
| ocu-miR-148a-5p | 1.61 | 0.0251 | ocu-miR-93-5p | −1.38 | 0.0238 |
| ocu-miR-532-3p | 1.52 | 0.0065 | ocu-miR-6529-5p | −1.37 | 0.0045 |
| ocu-miR-30b-5p | 1.50 | 0.0046 | ocu-miR-143-3p | −1.37 | 0.0104 |
| ocu-miR-31-5p | 1.40 | 0.0044 | ocu-miR-652-3p | −1.36 | 0.0241 |
| ocu-miR-214-5p | 1.37 | 0.0031 | ocu-miR-574-5p | −1.33 | 0.0123 |
| ocu-miR-222-3p | 1.37 | 0.0010 | ocu-miR-7a-5p | −1.32 | 0.0047 |
| ocu-miR-323a-3p | 1.34 | 0.0082 | ocu-miR-151-3p | −1.26 | 0.0102 |
| ocu-miR-502a-3p | 1.21 | 0.0126 | ocu-miR-15b-3p | −1.23 | 0.0097 |
| ocu-miR-130a-3p | 1.21 | 0.0185 | ocu-miR-671-5p | −1.21 | 0.0101 |
| ocu-miR-12092-3p | 1.20 | 0.0055 | ocu-let-7f-5p | −1.09 | 0.0268 |
| ocu-miR-29b-3p | 1.13 | 0.0128 | ocu-let-7a-5p | −1.08 | 0.0220 |
| ocu-miR-29a-3p | 1.10 | 0.0216 | ocu-miR-423-5p | −1.06 | 0.0259 |
| ocu-miR-335-5p | 1.07 | 0.0174 | ocu-miR-125b-3p | −1.06 | 0.0146 |
| ocu-miR-181b-5p | 1.07 | 0.0143 | ocu-miR-361-5p | −1.04 | 0.0240 |
| ocu-miR-128a-3p | −1.04 | 0.0161 | |||
| ocu-miR-128b-3p | −1.04 | 0.0161 |
| miRNA | log2(Fold Change) | p-Value | miRNA | log2(Fold Change) | p-Value |
|---|---|---|---|---|---|
| ocu-mir-708 * | 6.70 | 0.0000 | ocu-mir-574 * | −3.05 | 0.0007 |
| ocu-mir-134 * | 3.06 | 0.0001 | ocu-mir-503 * | −2.68 | 0.0000 |
| ocu-mir-125b-2 * | 2.65 | 0.0000 | ocu-mir-590 | −2.31 | 0.0133 |
| ocu-mir-140 * | 2.24 | 0.0000 | ocu-mir-30a * | −1.94 | 0.0001 |
| ocu-mir-29b-2 * | 2.04 | 0.0006 | ocu-mir-146a * | −1.93 | 0.0041 |
| ocu-mir-29b-1 * | 1.95 | 0.0011 | ocu-mir-671 * | −1.77 | 0.0014 |
| ocu-mir-214 * | 1.54 | 0.0003 | ocu-mir-181b-2 | −1.49 | 0.0112 |
| ocu-mir-26a | 1.35 | 0.0013 | ocu-mir-152 | −1.38 | 0.0106 |
| ocu-mir-23b | 1.05 | 0.0132 | ocu-mir-155 | −1.13 | 0.0104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesh, V.; Seol, D.; Gomez-Contreras, P.C.; Keen, H.L.; Shin, K.; Martin, J.A. Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration. Int. J. Mol. Sci. 2023, 24, 466. https://doi.org/10.3390/ijms24010466
Ganesh V, Seol D, Gomez-Contreras PC, Keen HL, Shin K, Martin JA. Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration. International Journal of Molecular Sciences. 2023; 24(1):466. https://doi.org/10.3390/ijms24010466
Chicago/Turabian StyleGanesh, Venkateswaran, Dongrim Seol, Piedad C. Gomez-Contreras, Henry L. Keen, Kyungsup Shin, and James A. Martin. 2023. "Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration" International Journal of Molecular Sciences 24, no. 1: 466. https://doi.org/10.3390/ijms24010466
APA StyleGanesh, V., Seol, D., Gomez-Contreras, P. C., Keen, H. L., Shin, K., & Martin, J. A. (2023). Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration. International Journal of Molecular Sciences, 24(1), 466. https://doi.org/10.3390/ijms24010466