Phenotypic and Safety Assessment of the Cheese Strain Lactiplantibacillus plantarum LL441, and Sequence Analysis of its Complete Genome and Plasmidome
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phenotypic Characterization of LL441
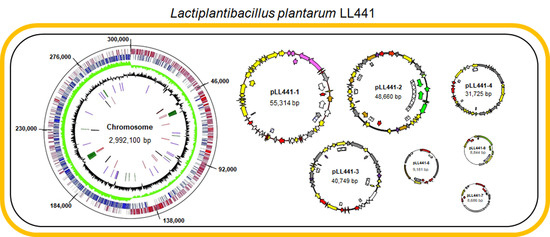
2.2. General Features of LL441 Genome
2.3. Plasmidome of LL441
2.4. Lactose Metabolism in LL441
2.5. Genetic Potential for Flavour Production
2.6. Genome Analysis and Other Functionalities
2.7. Genome Analysis and Safety
3. Materials and Methods
3.1. Culture Conditions
3.2. Phenotypic Characterization
3.3. Production of Volatile Compounds (VOCs) in Milk
3.4. Genome Sequencing
3.5. Genome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mayo, B.; Flórez, A.B. Lactic Acid Bacteria: Lactobacillus spp.: Lactobacillus plantarum. In Encyclopedia of Dairy Sciences; McSweeney, P.L.H., McNamara, J.P., Eds.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2022; Volume 4, pp. 206–217. [Google Scholar] [CrossRef]
- Papadopoulou, O.S.; Argyri, A.A.; Varzakis, E.E.; Tassou, C.C.; Chorianopoulos, N.G. Greek functional Feta cheese: Enhancing quality and safety using a Lactobacillus plantarum strain with probiotic potential. Food Microbiol. 2018, 74, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Speranza, B.; Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. A possible approach to assess acidification of meat starter cultures: A case study from some wild strains of Lactobacillus plantarum. J. Sci. Food Agric. 2017, 97, 2961–2968. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; Tzeneva, V.A.; Castioni, A.; Wels, M.; Phan, H.T.; Rademaker, J.L.; Starrenburg, M.J.; Kleerebezem, M.; Molenaar, D.; van Hylckama Vlieg, J.E. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010, 12, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef]
- Wu, C.; Huang, J.; Zhou, R. Genomics of lactic acid bacteria: Current status and potential applications. Crit. Rev. Microbiol. 2017, 43, 393–404. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef] [Green Version]
- Carpi, F.M.; Coman, M.M.; Silvi, S.; Picciolini, M.; Verdenelli, M.C.; Napolioni, V. Comprehensive pan-genome analysis of Lactiplantibacillus plantarum complete genomes. J. Appl. Microbiol. 2022, 132, 592–604. [Google Scholar] [CrossRef]
- Heuer, H.; Smalla, K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 2012, 36, 1083–1104. [Google Scholar] [CrossRef]
- Karim, A.S.; Curran, K.A.; Alper, H.S. Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res. 2013, 13, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Z.; Zhou, H. Identification of the conjugative and mobilizable plasmid fragments in the plasmidome using sequence signatures. Microb. Genom. 2020, 6, mgen000459. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Hu, T.; Qu, X.; Zhang, L.; Ding, Z.; Dong, A. Plasmids from food lactic acid bacteria: Diversity, similarity, and new developments. Int. J. Mol. Sci. 2015, 16, 13172–13202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLeod, A.; Fagerlund, A.; Rud, I.; Axelsson, L. Large plasmid complement resolved: Complete genome sequencing of Lactobacillus plantarum MF1298, a candidate probiotic strain associated with unfavorable effect. Microorganisms 2019, 7, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Mastrigt, O.; Di Stefano, E.; Hartono, S.; Abee, T.; Smid, E.J. Large plasmidome of dairy Lactococcus lactis subsp. lactis biovar diacetylactis FM03P encodes technological functions and appears highly unstable. BMC Genom. 2018, 19, 620. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; Renckens, B.; van Swam, I.; Peters, S.; van Kranenburg, R.; Kleerebezem, M.; de Vos, W.M. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl. Environ. Microbiol. 2005, 71, 8371–8382. [Google Scholar] [CrossRef] [Green Version]
- Settanni, L.; Moschetti, G. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 2010, 27, 691–697. [Google Scholar] [CrossRef]
- Mayo, B.; Rodríguez, J.; Vázquez, L.; Flórez, A.B. Microbial interactions within the cheese ecosystem and their application to improve quality and safety. Foods 2021, 10, 602. [Google Scholar] [CrossRef]
- Stefanovic, E.; Fitzgerald, G.; McAuliffe, O. Advances in the genomics and metabolomics of dairy lactobacilli: A review. Food Microbiol. 2017, 61, 33–49. [Google Scholar] [CrossRef]
- Mayo, B.; Hardisson, C.; Fernández-Braña, A. Selected characteristics of several strains of Lactobacillus plantarum. Microbiol. SEM 1989, 5, 105–122. [Google Scholar]
- González, B.; Arca, P.; Mayo, B.; Suárez, J.E. Detection, purification, and partial characterization of plantaricin C, a bacteriocin produced by a Lactobacillus plantarum strain of dairy origin. Appl. Environ. Microbiol. 1994, 60, 2158–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayo, B.; Arca, P.; González, B.; Suárez, J.E. Cloning and expression of the plasmid encoded β-galactosidase gene from a Lactobacillus plantarum strain of dairy origin. FEMS Microbiol. Lett. 1994, 122, 145–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flórez, A.B.; Mayo, B. Genome analysis of Lactobacillus plantarum LL441 and genetic characterisation of the locus for the lantibiotic plantaricin C. Front. Microbiol. 2018, 9, 1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado, S.; Mayo, B. Development of Lactobacillus plantarum LL441 and its plasmid-cured derivatives in cheese. J. Ind. Microbiol. Biotechnol. 2003, 30, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Safavi, F.; Rostami, A. Role of serine proteases in inflammation: Bowman-Birk protease inhibitor (BBI) as a potential therapy for autoimmune diseases. Exp. Mol. Pathol. 2013, 93, 428–433. [Google Scholar] [CrossRef]
- Dzutsev, A.; Goldszmid, R.S.; Viaud, S.; Zitvogel, L.; Trinchieri, G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur. J. Immunol. 2015, 45, 17–31. [Google Scholar] [CrossRef]
- Sakurama, H.; Kishino, S.; Uchibori, Y.; Yonejima, Y.; Ashida, H.; Kita, K.; Takahashi, S.; Ogawa, J. β-Glucuronidase from Lactobacillus brevis useful for baicalin hydrolysis belongs to glycoside hydrolase family 30. Appl. Microbiol. Biotechnol. 2014, 98, 4021–4032. [Google Scholar] [CrossRef]
- Flórez, A.B.; Egervärn, M.; Danielsen, M.; Tosi, L.; Morelli, L.; Lindgren, S.; Mayo, B. Susceptibility of Lactobacillus plantarum strains to six antibiotics and definition of new susceptibility-resistance cutoff values. Microb. Drug Resist. 2006, 12, 252–256. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2018, 85, e01738-18. [Google Scholar] [CrossRef] [Green Version]
- EFSA FEEDAP Panel. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef]
- Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; Del Río, B.; Álvarez, M.A. GABA-Producing Lactococcus lactis strains isolated from camel’s milk as starters for the production of GABA-enriched cheese. Foods 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Landaverde, P.A.; Velazquez, G.; Torres, J.A.; Qian, M.C. Quantitative determination of thermally derived off-flavor compounds in milk using solid-phase microextraction and gas chromatography. J. Dairy Sci. 2005, 88, 3764–3772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertuzzi, A.S.; McSweeney, P.L.H.; Rea, M.C.; Kilcawley, K.N. Detection of volatile compounds of cheese and their contribution to the flavor profile of surface-ripened cheese. Compr. Rev. Food Sci. Food Saf. 2018, 17, 371–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H. Volatile flavor compounds in yogurt: A review. Crit. Rev. Food Sci. Nutr. 2010, 50, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Settachaimongkon, S.; van Valenberg, H.J.; Gazi, I.; Nout, M.J.; van Hooijdonk, T.C.; Zwietering, M.H.; Smid, E.J. Influence of Lactobacillus plantarum WCFS1 on post-acidification, metabolite formation and survival of starter bacteria in set-yoghurt. Food Microbiol. 2016, 59, 14–22. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Rodríguez, J.; Vázquez, L.; Flórez, A.B.; Mayo, B. Phenotypic testing and genomic analysis of three lactic acid bacteria strains assembled as a consortium in a naturally fermented milk, and their metabolic interactions. Front. Microbiol. 2022, 13, 1000683. [Google Scholar] [CrossRef]
- Flórez, A.B.; Mayo, B. The plasmid complement of the cheese isolate Lactococcus garvieae IPLA 31405 revealed adaptation to the dairy environment. PLoS One 2015, 10, e0126101. [Google Scholar] [CrossRef]
- Flórez, A.B.; Vázquez, L.; Rodríguez, J.; Mayo, B. Direct recovery and molecular characterization of antibiotic resistance plasmids from cheese bacteria. Int. J. Mol. Sci. 2021, 22, 7801. [Google Scholar] [CrossRef]
- Kazi, T.A.; Acharya, A.; Mukhopadhyay, B.C.; Mandal, S.; Arukha, A.P.; Nayak, S.; Biswas, S.R. Plasmid-based gene expression systems for lactic acid bacteria: A review. Microorganisms 2022, 10, 1132. [Google Scholar] [CrossRef]
- Shmakov, S.A.; Sitnik, V.; Makarova, K.S.; Wolf, Y.I.; Severinov, K.V.; Koonin, E.V. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. MBio 2017, 8, 01307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopin, M.C.; Chopin, A.; Bidnenko, E. Phage abortive infection in lactococci: Variations on a theme. Curr. Opin. Microbiol. 2005, 8, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Prévots, F.; Daloyau, M.; Bonin, O.; Dumont, X.; Tolou, S. Cloning and sequencing of the novel abortive infection gene abiH of Lactococcus lactis ssp. lactis biovar. diacetylactis S94. FEMS Microbiol. Lett. 1996, 142, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, C.F.; Cailliez-Grimal, C.; Borges, F.; Revoll-Junelles, A.M. Review of lactose and galactose metabolism in lactic acid bacteria dedicated to expert genomic annotation. Trends Food Sci. Technol. 2019, 88, 121–132. [Google Scholar] [CrossRef]
- Fernández, M.; Margolles, A.; Suárez, J.E.; Mayo, B. Duplication of the β-galactosidase gene in some Lactobacillus plantarum strains. Int. J. Food Microbiol. 1999, 48, 113–123. [Google Scholar] [CrossRef]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [Green Version]
- Chambellon, E.; Yvon, M. CodY-regulated aminotransferases AraT and BcaT play a major role in the growth of Lactococcus lactis in milk by regulating the intracellular pool of amino acids. Appl. Environ. Microbiol. 2003, 69, 3061–3068. [Google Scholar] [CrossRef] [Green Version]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; and Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef]
- Smid, E.J.; Kleerebezem, M. Production of aroma compounds in lactic fermentations. Ann. Rev. Food Sci. Technol. 2014, 5, 313–326. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Smit, G.; Smit, B.A.; Engels, W.J. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, J.A.; Flórez, A.B.; Vázquez, L.; Vasek, O.M.; Mayo, B. Production of γ-aminobutyric acid (GABA) by lactic acid bacteria strains isolated from traditional, starter-free dairy products made of raw milk. Benef. Microbes 2019, 10, 579–587. [Google Scholar] [CrossRef]
- Yogeswara, I.B.A.; Maneerat, S.; Haltrich, D. Glutamate decarboxylase from lactic acid bacteria-A key enzyme in GABA synthesis. Microorganisms 2020, 8, 1923. [Google Scholar] [CrossRef] [PubMed]
- Kishino, S.; Ogawa, J.; Yokozeki, K.; Shimizu, S. Linoleic acid isomerase in Lactobacillus plantarum AKU1009a proved to be a multi-component enzyme system requiring oxidoreduction cofactors. Biosci. Biotechnol. Biochem. 2011, 75, 318–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishino, S.; Takeuchi, M.; Park, S.-B.; Ogawa, J. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. USA 2013, 110, 17808–17813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 2008, 72, 728–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, A.B.; Ashenden, A.; Köper, I. Model architectures for bacterial membranes. Biophys. Rev. 2022, 14, 111–143. [Google Scholar] [CrossRef]
- Takatsuka, Y.; Kmio, Y. Molecular dissection of the Selenomonas ruminantium cell envelope and lysine decarboxylase involved in the biosynthesis of a polyamine covalently linked to the cell wall peptidoglycan layer. Biosci. Biotechnol. Biochem. 2004, 68, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.; González-Guerra, A.; Vázquez, L.; Fernández-López, R.; Flórez, A.B.; de la Cruz, F.; Mayo, B. Isolation and phenotypic and genomic characterization of Tetragenococcus spp. from two Spanish traditional blue-veined cheeses made of raw milk. Int. J. Food Microbiol. 2022, 371, 109670. [Google Scholar] [CrossRef]
- Ladero, V.; Martín, M.C.; Mayo, B.; Flórez, A.B.; Fernández, M.; Álvarez, M.A. Genetic and functional analysis of the amine production capacity among starter and non-starter lactic acid bacteria isolated from artisanal cheeses. Eur. Food Res. Technol. 2015, 241, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Lim, J.M.; Kwon, S.J.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.D.; Jin, Q.; Chen, L.H.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Sayers, S.; Li, L.; Ong, E.; Deng, S.; Fu, G.; Lin, Y.; Yang, B.; Zhang, S.; Fa, Z.; Zhao, B.; et al. Victors: A web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acid Res. 2019, 47, D693–D700. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated identification, annotation, and classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef]



| Utilization of Carbohydrates | Enzyme Activity | Antibiotic Resistance | |||
|---|---|---|---|---|---|
| Carbohydrate | Degree of Utilization a | Enzyme | Activity b | Antibiotic | MIC c |
| D-ribose | ++++ | Alkaline phosphatase | 0 | Gentamicin | 1 |
| D-galactose | ++++ | Esterase (C 4) | <1 | Kanamycin | 16 |
| D-glucose | ++++ | Esterase lipase (C 8) | <1 | Streptomycin | 16 |
| D-fructose | ++++ | Lipase (C 14) | 0 | Neomycin | 1 |
| D-mannose | ++++ | Leucine arylamidase | >40 | Tetracycline | 32 |
| D-mannitol | ++++ | Valine arylamidase | 30 | Erythromycin | 0.25 |
| D-sorbitol | ++++ | Cystine arylamidase | <1 | Clindamycin | 1 |
| N-acetylglucosamine | +++ | Trypsin | 0 | Chloramphenicol | 8 |
| Amygdaline | ++ | α-chymotrypsin | 0 | Ampicillin | 1 |
| Arbutine | +++ | Acid phosphatase | 5 | Penicillin G | 4 |
| Esculine | ++++ | Naphthol-AS-BI-phosphohydrolase | 10 | Vancomycin | >256 |
| Salicine | +++ | α-galactosidase | 0 | Quinupristin-dalfopristin | 1 |
| D-cellobiose | ++++ | β-galactosidase | >40 | Linezolid | 4 |
| D-maltose | ++++ | β-glucuronidase | 0 | Trimethoprim | 0.25 |
| Lactose | ++++ | α-glucosidase | 20 | Ciprofloxacin | 16 |
| Mellibiose | ++++ | β-glucosidase | 30 | Rifampicin | 2 |
| D-sucrose | ++++ | N-acetyl-β-glucosaminidase | 30 | ||
| D-threhalose | ++ | α-mannosidase | 0 | ||
| Gentiobiose | ++ | α-fucosidase | 0 | ||
| D-turanose | ++ | ||||
| Compound | Control Unfermented Milk | Milk Fermented by LL441 |
|---|---|---|
| Acids | ||
| Acetic acid | - | 2.64 ± 0.39 |
| Butyric acid (butanoic acid) | - | 5.54 ± 1.51 |
| Caproic acid (hexanoic acid) | 6.58 | 50.45 ± 16.62 |
| Caprylic acid (octanoic acid) | 13.61 | 83.93 ± 24.54 |
| Perlargonic acid (nonanoic acid) | - | 1.66 ± 0.36 |
| Capric acid (n-decanoic acid) | 22.06 | 65.87 ± 14.71 |
| Caproleic acid (9-decenoic acid) | - | 5.06 ± 1.57 |
| Lauric acid (dodecanoic acid) | 4.11 | 9.56 ± 2.17 |
| Ketones | ||
| 2-Heptanone | 10.55 | 9.21 ± 5.32 |
| 2-Dodecanonea | - | 3.73 ± 2.15 |
| 2-Undecanone | 4.14 | 5.47 ± 1.35 |
| 2-Tridecanone | - | 2.61 ± 0.70 |
| 2-Nonanone | 8.74 | 9.43 ± 1.95 |
| 4-Methyl-2-hexanone a | 3.23 | 14.30 ± 8.25 |
| 6-pentyl-2H-pyran-2-one | 2.17 | 3.44 ± 0.53 |
| Lactones | ||
| γ-Dodecalactone | 1.78 | 1.69 ± 0.23 |
| δ-Dodecalactone | 1.95 | 2.52 ± 0.35 |
| Other | ||
| 1,3-bis(1,1-Dimethylethyl)benzene (sulfur compound) | - | 2.29 ± 1.33 |
| 4-Piperidinepropanoic acid, 1-benzoyl-3-(2-chloroethyl) a (alcohol) | - | 1.43 ± 0.42 |
| Tridecyl alcohol tri(oxyethylene) ethanol a (alcohol) | - | 2.66 ± 0.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flórez, A.B.; Vázquez, L.; Rodríguez, J.; Mayo, B. Phenotypic and Safety Assessment of the Cheese Strain Lactiplantibacillus plantarum LL441, and Sequence Analysis of its Complete Genome and Plasmidome. Int. J. Mol. Sci. 2023, 24, 605. https://doi.org/10.3390/ijms24010605
Flórez AB, Vázquez L, Rodríguez J, Mayo B. Phenotypic and Safety Assessment of the Cheese Strain Lactiplantibacillus plantarum LL441, and Sequence Analysis of its Complete Genome and Plasmidome. International Journal of Molecular Sciences. 2023; 24(1):605. https://doi.org/10.3390/ijms24010605
Chicago/Turabian StyleFlórez, Ana Belén, Lucía Vázquez, Javier Rodríguez, and Baltasar Mayo. 2023. "Phenotypic and Safety Assessment of the Cheese Strain Lactiplantibacillus plantarum LL441, and Sequence Analysis of its Complete Genome and Plasmidome" International Journal of Molecular Sciences 24, no. 1: 605. https://doi.org/10.3390/ijms24010605
APA StyleFlórez, A. B., Vázquez, L., Rodríguez, J., & Mayo, B. (2023). Phenotypic and Safety Assessment of the Cheese Strain Lactiplantibacillus plantarum LL441, and Sequence Analysis of its Complete Genome and Plasmidome. International Journal of Molecular Sciences, 24(1), 605. https://doi.org/10.3390/ijms24010605