TCGA RNA-Seq and Tumor-Infiltrating Lymphocyte Imaging Data Reveal Cold Tumor Signatures of Invasive Ductal Carcinomas and Estrogen Receptor-Positive Human Breast Tumors
Abstract
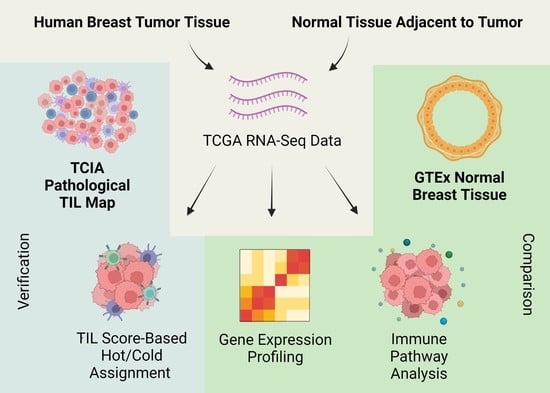
:1. Introduction
2. Results
2.1. Grouping Breast Tumor Samples into Hot and Cold Groups Based on Lymphocyte Scores
2.2. Cold NAT and Hot NAT Are Immunologically Active but Cold Tumors Are Immunologically Inactive
2.3. High M2 Macrophages in Cold Tumors
2.4. Genes Correlated with Low Lymphocyte Score in Cold Tumors Reveal Low Antigen Presentation and Increased Matrix Remodeling
2.5. Infiltrating Ductal Carcinoma and ER-Positive Tumors Exhibit Cold Tumor Signiatures
2.6. RNA-Seq-Based Hot/Cold Classification and Pathological TIL Patterns Mostly Coincide
3. Discussion
4. Materials and Methods
4.1. Expression Data of Breast Cancer, Normal Tissue Adjacent to Tumor (NAT), and Normal Healthy Tissue Samples
4.2. Immune Cell Analysis and Hot/Cold Status Assignment
4.3. Differential Gene Expression Analysis
4.4. Pathway Analysis
4.5. Verification of RNA-Seq-Based Tumor Dichotomy Using Matched Pathology Images
4.6. Fisher’s Exact Test
4.7. Survival Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marra, A.; Viale, G.; Curigliano, G. Recent advances in triple negative breast cancer: The immunotherapy era. BMC Med. 2019, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Yi, M.; Jiao, D.; Xu, H.; Liu, Q.; Zhao, W.; Han, X.; Wu, K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef]
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold tumors: A therapeutic challenge for immunotherapy. Front. Immunol. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Bilusic, M.; Gulley, J.L. Local immunotherapy: A way to convert tumors from “Cold” to “Hot”. JNCI J. Natl. Cancer Inst. 2017, 109, djx132. [Google Scholar] [CrossRef]
- Marrogi, A.J.; Munshi, A.; Merogi, A.J.; Ohadike, Y.; El-Habashi, A.; Marrogi, O.L.; Freeman, S.M. Study of tumor infiltrating lymphocytes and transforming growth factor-β as prognostic factors in breast carcinoma. Int. J. Cancer 1997, 74, 492–501. [Google Scholar] [CrossRef]
- Naito, Y.; Saito, K.; Shiiba, K.; Ohuchi, A.; Saigenji, K.; Nagura, H.; Ohtani, H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998, 58, 3491–3494. [Google Scholar]
- Nakano, O.; Sato, M.; Naito, Y.; Suzuki, K.; Orikasa, S.; Aizawa, M.; Suzuki, Y.; Shintaku, I.; Nagura, H.; Ohtani, H. Proliferative activity of intratumoral CD8+ T-lymphocytes as a prognostic factor in human renal cell carcinoma: Clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001, 61, 5132–5136. [Google Scholar]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef]
- Kruger, S.; Ilmer, M.; Kobold, S.; Cadilha, B.L.; Endres, S.; Ormanns, S.; Schuebbe, G.; Renz, B.W.; D’Haese, J.G.; Schloesser, H. Advances in cancer immunotherapy 2019–latest trends. J. Exp. Clin. Cancer Res. 2019, 38, 1–11. [Google Scholar] [CrossRef]
- Bou-Dargham, M.J.; Liu, Y.; Sang, Q.-X.A.; Zhang, J. Subgrouping breast cancer patients based on immune evasion mechanisms unravels a high involvement of transforming growth factor-beta and decoy receptor 3. PLoS ONE 2018, 13, e0207799. [Google Scholar] [CrossRef]
- Haanen, J.B. Converting cold into hot tumors by combining immunotherapies. Cell 2017, 170, 1055–1056. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Lin, Y.-C.; Yao, P.-L.; Yuan, A.; Chen, H.-Y.; Shun, C.-T.; Tsai, M.-F.; Chen, C.-H.; Yang, P.-C. Tumor-associated macrophages: The double-edged sword in cancer progression. J. Clin. Oncol. 2005, 23, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Deng, G. Tumor-infiltrating regulatory T cells: Origins and features. Am. J. Clin. Exp. Immunol. 2018, 7, 81. [Google Scholar] [PubMed]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.D.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T cells exhibit distinct features in human breast cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef]
- Vinay, D.S.; Ryan, E.P.; Pawelec, G.; Talib, W.H.; Stagg, J.; Elkord, E.; Lichtor, T.; Decker, W.K.; Whelan, R.L.; Kumara, H.M.C.S.; et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015, 35, 185. [Google Scholar]
- Corthay, A. How do regulatory T cells work? Scand. J. Immunol. 2009, 70, 326–336. [Google Scholar] [CrossRef]
- Shevach, E.M.; Tran, D.Q.; Davidson, T.S.; Andersson, J. The critical contribution of TGF-β to the induction of Foxp3 expression and regulatory T cell function. Eur. J. Immunol. 2008, 38, 915–917. [Google Scholar] [CrossRef]
- Arnold, K.M.; Opdenaker, L.M.; Flynn, D.; Sims-Mourtada, J. Wound healing and cancer stem cells: Inflammation as a driver of treatment resistance in breast cancer. Cancer Growth Metastasis 2015, 8, CGM.S11286. [Google Scholar] [CrossRef]
- Liao, Z.; Chua, D.; Tan, N.S. Reactive oxygen species: A volatile driver of field cancerization and metastasis. Mol. Cancer 2019, 18, 1–10. [Google Scholar] [CrossRef]
- Murdoch, C.; Muthana, M.; Coffelt, S.B.; Lewis, C.E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer 2008, 8, 618–631. [Google Scholar] [CrossRef]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef]
- De Guillebon, E.; Dardenne, A.; Saldmann, A.; Séguier, S.; Tran, T.; Paolini, L.; Lebbe, C.; Tartour, E. Beyond the concept of cold and hot tumors for the development of novel predictive biomarkers and the rational design of immunotherapy combination. Int. J. Cancer 2020, 147, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]
- Gide, T.N.; Quek, C.; Menzies, A.M.; Tasker, A.T.; Shang, P.; Holst, J.; Madore, J.; Lim, S.Y.; Velickovic, R.; Wongchenko, M. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 2019, 35, 238–255.e236. [Google Scholar] [CrossRef]
- Xiong, X.; Chen, C.; Li, X.; Yang, J.; Zhang, W.; Wang, X.; Zhang, H.; Peng, M.; Li, L.; Luo, P. Identification of a novel defined inflammation-related long noncoding RNA signature contributes to predicting prognosis and distinction between the cold and hot tumors in bladder cancer. Front. Oncol. 2023, 13, 972558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Zhou, X.; Fang, D.; Ou, X.; Ye, J.; Peng, J.; Xu, J. Necroptosis-related lncRNAs: Predicting prognosis and the distinction between the cold and hot tumors in gastric cancer. J. Oncol. 2021, 2021, 6718443. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, L.; Wang, Y.; Zuo, Y.; Chen, D.; Guo, X. Comprehensive prediction of immune microenvironment and hot and cold tumor differentiation in cutaneous melanoma based on necroptosis-related lncRNA. Sci. Rep. 2023, 13, 7299. [Google Scholar] [CrossRef]
- Du, J.-w.; Li, G.-q.; Li, Y.-s.; Qiu, X.-g. Identification of prognostic biomarkers related to the tumor microenvironment in thyroid carcinoma. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Institute, N.C. The Cancer Genome Atlas. Available online: https://www.cancer.gov (accessed on 3 February 2023).
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Yang, T.-H.O.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A. The immune landscape of cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef] [PubMed]
- Carithers, L.J.; Ardlie, K.; Barcus, M.; Branton, P.A.; Britton, A.; Buia, S.A.; Compton, C.C.; DeLuca, D.S.; Peter-Demchok, J.; Gelfand, E.T. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreservation Biobanking 2015, 13, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Tumor-Infiltrating Lymphocytes Maps from TCGA H&E Whole Slide Pathology Images [Data set]. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018, 23, 181–193.e187. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Pénault-Llorca, F. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Vonderheide, R.H.; Domchek, S.M.; Clark, A.S. Immunotherapy for breast cancer: What are we missing? Clin. Cancer Res. 2017, 23, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Desmedt, C.; Salgado, R.; Fornili, M.; Pruneri, G.; Van den Eynden, G.; Zoppoli, G.; Rothe, F.; Buisseret, L.; Garaud, S.; Willard-Gallo, K. Immune infiltration in invasive lobular breast cancer. JNCI J. Natl. Cancer Inst. 2018, 110, 768–776. [Google Scholar] [CrossRef]
- Richard, F.; Majjaj, S.; Venet, D.; Rothé, F.; Pingitore, J.; Boeckx, B.; Marchio, C.; Clatot, F.; Bertucci, F.; Mariani, O. Characterization of Stromal Tumor-infiltrating Lymphocytes and Genomic Alterations in Metastatic Lobular Breast Cancer. Clin. Cancer Res. 2020, 26, 6254–6265. [Google Scholar] [CrossRef]
- Solinas, C.; Carbognin, L.; De Silva, P.; Criscitiello, C.; Lambertini, M. Tumor-infiltrating lymphocytes in breast cancer according to tumor subtype: Current state of the art. Breast 2017, 35, 142–150. [Google Scholar] [CrossRef]
- Annaratone, L.; Cascardi, E.; Vissio, E.; Sarotto, I.; Chmielik, E.; Sapino, A.; Berrino, E.; Marchiò, C. The multifaceted nature of tumor microenvironment in breast carcinomas. Pathobiology 2020, 87, 125–142. [Google Scholar] [CrossRef]
- Nguyen, B.; Veys, I.; Leduc, S.; Bareche, Y.; Majjaj, S.; Brown, D.N.; Boeckx, B.; Lambrechts, D.; Sotiriou, C.; Larsimont, D. Genomic, transcriptomic, epigenetic, and immune profiling of mucinous breast cancer. JNCI J. Natl. Cancer Inst. 2019, 111, 742–746. [Google Scholar] [CrossRef]
- Dieci, M.V.; Miglietta, F.; Guarneri, V. Immune infiltrates in breast cancer: Recent updates and clinical implications. Cells 2021, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Disis, M.L.; Stanton, S.E. Triple-negative breast cancer: Immune modulation as the new treatment paradigm. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e25–e30. [Google Scholar] [CrossRef]
- Nair, M.G.; VP, S.; Patil, S.; CE, A.; Mukherjee, G.; Kumar, R.V.; Prabhu, J.S.; TS, S. miR-18a Mediates Immune Evasion in ER-Positive Breast Cancer through Wnt Signaling. Cells 2022, 11, 1672. [Google Scholar] [CrossRef]
- Terranova-Barberio, M.; Pawlowska, N.; Dhawan, M.; Moasser, M.; Chien, A.J.; Melisko, M.E.; Rugo, H.; Rahimi, R.; Deal, T.; Daud, A. Exhausted T cell signature predicts immunotherapy response in ER-positive breast cancer. Nat. Commun. 2020, 11, 3584. [Google Scholar] [CrossRef]
- Kohler, B.A.; Sherman, R.L.; Howlader, N.; Jemal, A.; Ryerson, A.B.; Henry, K.A.; Boscoe, F.P.; Cronin, K.A.; Lake, A.; Noone, A.-M. Annual report to the nation on the status of cancer, 1975-2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J. Natl. Cancer Inst. 2015, 107, djv048. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.-C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S. Mechanisms of tumor escape in the context of the T-cell-inflamed and the non-T-cell-inflamed tumor microenvironment. Int. Immunol. 2016, 28, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Comito, G.; Giannoni, E.; Segura, C.; Barcellos-de-Souza, P.; Raspollini, M.; Baroni, G.; Lanciotti, M.; Serni, S.; Chiarugi, P. Cancer-associated fibroblasts and M2-polarized macrophages synergize during prostate carcinoma progression. Oncogene 2014, 33, 2423–2431. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Mukherjee, S.; Choudhury, S.; Gupta, P.; Adhikary, A.; Baral, R.; Chattopadhyay, S. Reactive oxygen species in the tumor niche triggers altered activation of macrophages and immunosuppression: Role of fluoxetine. Cell. Signal. 2015, 27, 1398–1412. [Google Scholar] [CrossRef]
- Hashimoto, O.; Yoshida, M.; Koma, Y.i.; Yanai, T.; Hasegawa, D.; Kosaka, Y.; Nishimura, N.; Yokozaki, H. Collaboration of cancer-associated fibroblasts and tumour-associated macrophages for neuroblastoma development. J. Pathol. 2016, 240, 211–223. [Google Scholar] [CrossRef]
- Ademmer, K.; Ebert, M.; Müller-Ostermeyer, F.; Friess, H.; Büchler, M.; Schubert, W.; Malfertheiner, P. Effector T lymphocyte subsets in human pancreatic cancer: Detection of CD8+ CD18+ cells and CD8+ CD103+ cells by multi-epitope imaging. Clin. Exp. Immunol. 1998, 112, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Beatty, G.L.; Winograd, R.; Evans, R.A.; Long, K.B.; Luque, S.L.; Lee, J.W.; Clendenin, C.; Gladney, W.L.; Knoblock, D.M.; Guirnalda, P.D. Exclusion of T cells from pancreatic carcinomas in mice is regulated by Ly6Clow F4/80+ extratumoral macrophages. Gastroenterology 2015, 149, 201–210. [Google Scholar] [CrossRef]
- Jackute, J.; Zemaitis, M.; Pranys, D.; Sitkauskiene, B.; Miliauskas, S.; Vaitkiene, S.; Sakalauskas, R. Distribution of M1 and M2 macrophages in tumor islets and stroma in relation to prognosis of non-small cell lung cancer. BMC Immunol. 2018, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Griffith, J.K.; Bisoffi, M. Mammary field cancerization: Molecular evidence and clinical importance. Breast Cancer Res. Treat. 2009, 118, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Curtius, K.; Wright, N.A.; Graham, T.A. An evolutionary perspective on field cancerization. Nat. Rev. Cancer 2018, 18, 19–32. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Degnim, A.C.; Brahmbhatt, R.D.; Radisky, D.C.; Hoskin, T.L.; Stallings-Mann, M.; Laudenschlager, M.; Mansfield, A.; Frost, M.H.; Murphy, L.; Knutson, K. Immune cell quantitation in normal breast tissue lobules with and without lobulitis. Breast Cancer Res. Treat. 2014, 144, 539–549. [Google Scholar] [CrossRef]
- Gratz, I.K.; Campbell, D.J. Organ-specific and memory treg cells: Specificity, development, function, and maintenance. Front. Immunol. 2014, 5, 333. [Google Scholar] [CrossRef]
- Lança, T.; Silva-Santos, B. The split nature of tumor-infiltrating leukocytes: Implications for cancer surveillance and immunotherapy. Oncoimmunology 2012, 1, 717–725. [Google Scholar] [CrossRef]
- Pauken, K.E.; Wherry, E.J. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015, 36, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A. Adaptive Immune Resistance: How Cancer Protects from Immune AttackAdaptive Immune Resistance. Cancer Discov. 2015, 5, 915–919. [Google Scholar] [CrossRef]
- Escors, D. Tumour immunogenicity, antigen presentation, and immunological barriers in cancer immunotherapy. New J. Sci. 2014, 2014, 734515. [Google Scholar] [CrossRef]
- Kalos, M.; June, C.H. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 2013, 39, 49–60. [Google Scholar] [CrossRef]
- Makkouk, A.; Weiner, G.J. Cancer Immunotherapy and Breaking Immune Tolerance: New Approaches to an Old ChallengeCancer Immunotherapy and Breaking Immune Tolerance. Cancer Res. 2015, 75, 5–10. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. In Wiley Interdisciplinary Reviews: Computational Statistics 3.2; Wiley Online Library: Hoboken, NJ, USA, 2011. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 1–17. [Google Scholar] [CrossRef]
- Giles, J.R.; Ngiow, S.F.; Manne, S.; Baxter, A.E.; Khan, O.; Wang, P.; Staupe, R.; Abdel-Hakeem, M.S.; Huang, H.; Mathew, D. Shared and distinct biological circuits in effector, memory and exhausted CD8+ T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat. Immunol. 2022, 23, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Zander, R.; Schauder, D.; Xin, G.; Nguyen, C.; Wu, X.; Zajac, A.; Cui, W. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 2019, 51, 1028–1042.e1024. [Google Scholar] [CrossRef] [PubMed]






| Histology/Receptor Status | Cold Samples | Hot Samples | Total Samples | p Value * |
|---|---|---|---|---|
| Infiltrating Lobular Carcinoma | 89 (44.50%) | 111 (55.50%) | 200 | 6.34 × 10−4 |
| Infiltrating Ductal Carcinoma | 462 (60%) | 308 (40.00%) | 770 | 1.68 × 10−2 |
| Mucinous Carcinoma | 16 (94.12%) | 1 (5.88%) | 17 | 1.91 × 10−3 |
| ER-positive | 475 (59.97%) | 317 (40.03%) | 792 | 1.23 × 10−2 |
| ER-negative | 119 (50.64%) | 116 (49.36%) | 235 | |
| HER2-positive | 97 (60.25%) | 64 (39.75%) | 161 | 7.85 × 10−1 |
| HER2-negative | 324 (58.80%) | 227 (41.20%) | 551 | |
| TNBC | 61 (54.46%) | 51 (45.54%) | 112 | 4.42 × 10−1 |
| Histology/Receptor Status | Cold Samples a | Hot Samples b | Total Samples | p Value * |
|---|---|---|---|---|
| Lobular Carcinoma | 87 (74.36%) | 30 (25.64%) | 117 | 2.91 × 10−3 |
| Infiltrating Ductal Carcinoma | 239 (58.01%) | 173 (41.99%) | 412 | 7.65 × 10−4 |
| Infiltrating Ductal and Lobular Carcinoma | 12 (54.55%) | 10 (45.45%) | 22 | 5.02 × 10−1 |
| Mucinous Carcinoma | 8 (88.89%) | 1 (11.11%) | 9 | 1.64 × 10−1 |
| ER-positive | 318 (67.52%) | 153 (32.48%) | 471 | 1.97 × 10−6 |
| ER-negative | 57 (44.19%) | 72 (55.81%) | 129 | |
| HER2-positive | 52 (58.43%) | 37 (41.57%) | 89 | 4.08 × 10−1 |
| HER2-negative | 323 (63.21%) | 188 (36.79%) | 511 | |
| TNBC | 40 (40.40%) | 59 (59.60%) | 99 | 1.31 × 10−6 |
| Histology/Receptor Status | RNA-Seq | TIL Map |
|---|---|---|
| Infiltrating Lobular Carcinoma/Lobular Carcinoma | Significant (Cold 44.50%, Hot 55.50%) | Significant (Cold 74.36%, Hot 25.64%) |
| Infiltrating Ductal Carcinoma | Significant (Cold 60%, Hot 40%) | Significant (Cold 58.01%, Hot 41.99%) |
| Mucinous Carcinoma | Significant (Cold 94.12%, Hot 5.88%) | Non-Significant (Cold 88.89%, Hot 11.11%) |
| ER | Significant (pos.: Cold 59.97%, Hot 40.03%) (neg.: Cold 50.64%, Hot 49.36%) | Significant (pos.: Cold 67.52%, Hot 32.48%) (neg.: Cold 44.19%, Hot 55.81%) |
| HER2 | Non-Significant (pos.: Cold 60.25%, Hot 39.75%) (neg.: Cold 58.80%, Hot 41.20%) | Non-Significant (pos.: Cold 58.43%, Hot 41.57%) (neg.: Cold 63.21%, Hot 36.79%) |
| TNBC | Non-Significant (Cold 54.46%, Hot 45.54%) | Significant (Cold 40.40%, Hot 59.60%) |
| TCGA (1194) | TCGA Cancer Samples (1082) | Cold Tumor Samples (627) |
| Hot Tumor Samples (455) | ||
| TCGA NAT Samples (112) | Cold NAT Samples (62) | |
| Hot NAT Samples (50) | ||
| GTEx (115) | GTEx Normal (115) | GTEx Normal (115) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bou-Dargham, M.J.; Sha, L.; Sarker, D.B.; Krakora-Compagno, M.Z.; Chen, Z.; Zhang, J.; Sang, Q.-X.A. TCGA RNA-Seq and Tumor-Infiltrating Lymphocyte Imaging Data Reveal Cold Tumor Signatures of Invasive Ductal Carcinomas and Estrogen Receptor-Positive Human Breast Tumors. Int. J. Mol. Sci. 2023, 24, 9355. https://doi.org/10.3390/ijms24119355
Bou-Dargham MJ, Sha L, Sarker DB, Krakora-Compagno MZ, Chen Z, Zhang J, Sang Q-XA. TCGA RNA-Seq and Tumor-Infiltrating Lymphocyte Imaging Data Reveal Cold Tumor Signatures of Invasive Ductal Carcinomas and Estrogen Receptor-Positive Human Breast Tumors. International Journal of Molecular Sciences. 2023; 24(11):9355. https://doi.org/10.3390/ijms24119355
Chicago/Turabian StyleBou-Dargham, Mayassa J., Linlin Sha, Drishty B. Sarker, Martina Z. Krakora-Compagno, Zhui Chen, Jinfeng Zhang, and Qing-Xiang Amy Sang. 2023. "TCGA RNA-Seq and Tumor-Infiltrating Lymphocyte Imaging Data Reveal Cold Tumor Signatures of Invasive Ductal Carcinomas and Estrogen Receptor-Positive Human Breast Tumors" International Journal of Molecular Sciences 24, no. 11: 9355. https://doi.org/10.3390/ijms24119355
APA StyleBou-Dargham, M. J., Sha, L., Sarker, D. B., Krakora-Compagno, M. Z., Chen, Z., Zhang, J., & Sang, Q. -X. A. (2023). TCGA RNA-Seq and Tumor-Infiltrating Lymphocyte Imaging Data Reveal Cold Tumor Signatures of Invasive Ductal Carcinomas and Estrogen Receptor-Positive Human Breast Tumors. International Journal of Molecular Sciences, 24(11), 9355. https://doi.org/10.3390/ijms24119355