Biology of Two-Spotted Spider Mite (Tetranychus urticae): Ultrastructure, Photosynthesis, Guanine Transcriptomics, Carotenoids and Chlorophylls Metabolism, and Decoyinine as a Potential Acaricide
Abstract
:1. Introduction
2. Results
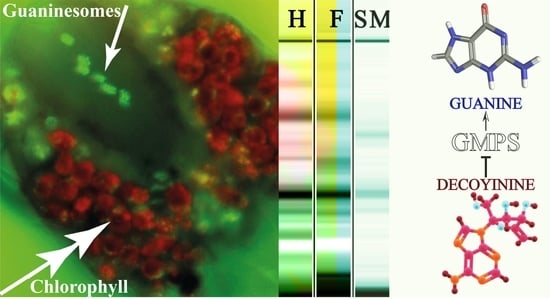
2.1. Fluorescence Microscopy Differentiates Chlorophyll and Guanine Metabolism in the TSSM
2.2. TSSM Ultrastructure Reveals the Presence of Intact Chloroplasts
2.3. Guanine Is Excreted by TSSM
2.4. Guanine Is Produced by TSSMs through the Upregulation of Most of the Guanine Biosynthesis Coding Genes
2.5. Decoyinine Inhibits GMPS and Reduces the TSSM Damage to Lima Bean Leaves
2.6. TSSMs Contain Mainly Pheophytin, and Their Feeding Activity Decreases Chlorophyll and Increases Pheophytin Contents in Lima Bean Leaves
2.7. TSSMs Contain a High Content of Carotenoids, and Their Feeding Activity Decreases the Total Carotenoid Content of Lima Bean Leaves
2.8. TSSMs Feeding Activity Alters the Protein and Sugar Content of Lima Bean Leaves
2.9. Do TSSMs Retain Photosynthetic Activity?
2.10. TSSMs Contain a Low Amount of Ribulose.1,5-Bisphosphate Carboxylase Oxygenase (RubisCO) Protein and Lack RubisCO Enzyme Activity
3. Discussion
4. Materials and Methods
4.1. Mite Strains and Rearing
4.2. Epifluorescence Microscopy
4.3. Confocal Laser Scanning Microscopy (CLSM)
4.4. Transmission Electron Microscopy
4.5. RNA Preparation, cDNA Cloning, and qRT-PCR Assays
4.6. Acaricidal Activity of Decoyinine (DCY)
4.7. Chlorophyll and Carotenoid Extraction
4.8. Liquid Chromatography of Chlorophyll, Chlorophyll Degradation Products, and Carotenoids
4.9. Liquid Chromatography and Tandem Mass Spectrometry and Guanine
4.10. Photosynthetic Activity in TSSMs
4.11. Total Sugar Content of Lima Bean Leaves and TSSMs
4.12. Total Protein Content of Lima Bean Leaves and TSSMs
4.13. Capillary Gel Electrophoresis
4.14. RubisCO Extraction and Enzyme Activity
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Leeuwen, T.; Vontas, J.; Tsagkarakou, A.; Dermauw, W.; Tirry, L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem. Mol. Biol. 2010, 40, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Helle, H.; Sabelis, M.W. Spider Mites. Their Biology, Natural Enemies and Control; Elsevier: Amsterdam, The Netherlands, 1985; Volume 1A. [Google Scholar]
- Iida, J.; Desaki, Y.; Hata, K.; Uemura, T.; Yasuno, A.; Islam, M.; Maffei, M.E.; Ozawa, R.; Nakajima, T.; Galis, I.; et al. Tetranins: New putative spider mite elicitors of host plant defense. New Phytol. 2019, 224, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Esteves Filho, A.B.; de Oliveira, J.V.; Torres, J.B.; Gondim, M.G. Compared Biology and behavior of Tetranychus urticae Koch (Acari: Tetranychidae) and Phytoseiulus macropilis (Banks) (Acari: Phytoseiidae) on Bollgard (TM) and non-transgenic isoline cotton. Neotrop. Entomol. 2010, 39, 338–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Occhipinti, A.; Maffei, M.E. Chlorophyll and its degradation products in the two-spotted spider mite, Tetranychus urticae: Observations using epifluorescence and confocal laser scanning microscopy. Exp. Appl. Acarol. 2013, 61, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Khajehali, J.; Van Nieuwenhuyse, P.; Demaeght, P.; Tirry, L.; Van Leeuwen, T. Acaricide resistance and resistance mechanisms in Tetranychus urticae populations from rose greenhouses in the Netherlands. Pest Manag. Sci. 2011, 67, 1424–1433. [Google Scholar] [CrossRef]
- Bensoussan, N.; Zhurov, V.; Yamakawa, S.; O’Neil, C.H.; Suzuki, T.; Grbic, M.; Grbic, V. The digestive system of the Two-Spotted Spider Mite, Tetranychus urticae Koch, in the context of the mite-plant interaction. Front. Plant Sci. 2018, 9, 18. [Google Scholar] [CrossRef] [Green Version]
- Pinsk, N.; Wagner, A.; Cohen, L.; Smalley, C.J.H.; Hughes, C.E.; Zhang, G.; Pavan, M.J.; Casati, N.; Jantschke, A.; Goobes, G.; et al. Biogenic guanine crystals are solid solutions of guanine and other purine metabolites. J. Am. Chem. Soc. 2022, 144, 5180–5189. [Google Scholar] [CrossRef]
- McEnroe, W.D. Guanine Excretion by the Two-Spotted Spider Mite (Tetranychus telarius L.). Ann. Entomol. Soc. Am. 1961, 54, 925–926. [Google Scholar] [CrossRef]
- Wagner, A.; Ezersky, V.; Maria, R.; Upcher, A.; Lemcoff, T.; Aflalo, E.D.; Lubin, Y.; Palmer, B.A. The non-classical crystallization mechanism of a composite biogenic guanine crystal. Adv. Mater. 2022, 34, 10. [Google Scholar] [CrossRef]
- Wiesmann, R. Untersuchungen über die Verdauungsvorgänge bei der gemeinen Spinnmilbe, Tetranychus urticae Koch. Z. Angew. Entomol. 1968, 61, 457–465. [Google Scholar] [CrossRef]
- Van der Geest, L.P. Aspects of physiology. In Spider Mites. Their Biology, Natural Enemies and Control; Helle, H., Sabelis, M.W., Eds.; Elsevier: Amsterdam, The Netherlands, 1985; Volume 1A, pp. 171–184. [Google Scholar]
- Van Leeuwen, T.; Vanholme, B.; Van Pottelberge, S.; Van Nieuwenhuyse, P.; Nauen, R.; Tirry, L.; Denholm, I. Mitochondrial heteroplasmy and the evolution of insecticide resistance: Non-Mendelian inheritance in action. Proc. Natl. Acad. Sci. USA 2008, 105, 5980–5985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirello, P.; Pozzebon, A.; Cassanelli, S.; Van Leeuwen, T.; Duso, C. Resistance to acaricides in Italian strains of Tetranychus urticae: Toxicological and enzymatic assays. Exp. Appl. Acarol. 2012, 57, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Mitani, T.; Heinze, J.E.; Freese, E. Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochem. Biophys. Res. Commun. 1977, 77, 1118–1125. [Google Scholar] [CrossRef]
- Yu, L.; Zhou, W.; She, Y.; Ma, H.; Cai, Y.S.; Jiang, M.; Deng, Z.; Price, N.P.J.; Chen, W. Efficient biosynthesis of nucleoside cytokinin angustmycin A containing an unusual sugar system. Nat. Commun. 2021, 12, 6633. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Shi, J.H.; Fan, T.; Wang, C.; Liu, L.; Jin, H.; Foba, C.N.; Wang, M.Q. The control of the brown planthopper by the rice Bph14 gene is affected by nitrogen. Pest Manag. Sci. 2020, 76, 3649–3656. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, Y.Y.; Gui, W.; Zhang, Q.X.; Xu, G.; Yang, G.Q. Priming of rice seed with decoyinine enhances resistance against the brown planthopper Nilparvata lugens. Crop Prot. 2022, 157, 8. [Google Scholar] [CrossRef]
- Shah, A.Z.; Ma, C.; Zhang, Y.Y.; Zhang, Q.X.; Xu, G.; Yang, G.Q. Decoyinine induced resistance in rice against small brown planthopper Laodelphax striatellus. Insects 2022, 13, 15. [Google Scholar] [CrossRef]
- Pumilia, G.; Cichon, M.J.; Cooperstone, J.L.; Giuffrida, D.; Dugo, G.; Schwartz, S.J. Changes in chlorophylls, chlorophyll degradation products and lutein in pistachio kernels (Pistacia vera L.) during roasting. Food Res. Int. 2014, 65, 193–198. [Google Scholar] [CrossRef]
- Bate, N.J.; Rothstein, S.J.; Thompson, J.E. Expression of nuclear and chloroplast photosynthesis-specific genes during leaf senescence. J. Exp. Bot. 1991, 42, 801–811. [Google Scholar] [CrossRef]
- Bondada, B.R.; Oosterhuis, D.M.; Tugwell, N.P.; Kim, K.S. Physiological, and cytological studies of two Spotted Spider-Mite, Tetranychus urticae K (Acari, Tetranychidae) injury in cotton. Southw. Entomol. 1995, 20, 171–180. [Google Scholar]
- Fathipour, Y.; Maleknia, B.; Bagheri, A.; Soufbaf, M.; Zalucki, M.P. Spider mite host plant resistance traits improve the predatory performance of Phytoseiulus persimilis on cucumber, despite negative life history impacts. Biol. Control 2019, 138, 9. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.; Goswami, M.T.; Pathi, S.S.; Dodson, M.; Chandrashekar, D.S.; Agarwal, S.; Nepal, S.; Hodigere Balasubramanya, S.A.; Siddiqui, J.; Lonigro, R.J.; et al. Expression and role of PAICS, a de novo purine biosynthetic gene in prostate cancer. Prostate 2017, 77, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Kar, B.; Varshney, R.; Roy, P.; Sharma, A.K. Characterization of AICAR transformylase/IMP cyclohydrolase (ATIC) from Staphylococcus lugdunensis. FEBS J. 2017, 284, 4233–4261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tesmer, J.J.; Klem, T.J.; Deras, M.L.; Davisson, V.J.; Smith, J.L. The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat. Struct. Biol. 1996, 3, 74–86. [Google Scholar] [CrossRef]
- Rodriguez-Suarez, R.; Xu, D.M.; Veillette, K.; Davison, J.; Sillaots, S.; Kauffman, S.; Hu, W.Q.; Bowman, J.; Martel, N.; Trosok, S.; et al. Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem. Biol. 2007, 14, 1163–1175. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, J.; Lou, L. Biochemical characterization of human GMP synthetase. J. Biol. Chem. 1995, 270, 7347–7353. [Google Scholar] [CrossRef] [Green Version]
- Deras, M.L.; Chittur, S.V.; Davisson, V.J. N-2-hydroxyguanosine 5′-monophosphate is a time-dependent inhibitor of Escherichia coli guanosine monophosphate synthetase. Biochemistry 1999, 38, 303–310. [Google Scholar] [CrossRef]
- Yuntsen, H.; Yonehara, H.; Ui, H. Studies on a new antibiotic, angustmycin. J. Antibiot. 1954, 7, 113–115. [Google Scholar]
- Adesanya, A.W.; Lavine, M.D.; Moural, T.W.; Lavine, L.C.; Zhu, F.; Walsh, D.B. Mechanisms and management of acaricide resistance for Tetranychus urticae in agroecosystems. J. Pest Sci. 2021, 94, 639–663. [Google Scholar] [CrossRef]
- Chen, Q.; Liang, X.; Wu, C.L.; Gao, J.T.; Chen, Q.; Zhang, Z. Density threshold-based acaricide application for the two-spotted spider mite Tetranychus urticae on cassava: From laboratory to the field. Pest Manag. Sci. 2019, 75, 2634–2641. [Google Scholar] [CrossRef]
- Born, F.D.; da Camara, C.A.G.; de Moraes, M.M.; Melo, J. Acaricidal properties of the selected components, blends and essential oils of species of genus Protium (Burseraceae) against Tetranychus urticae (Acari: Tetranychidae). Austral. Entomol. 2022, 61, 258–264. [Google Scholar] [CrossRef]
- Alpkent, Y.N.; Inak, E.; Ulusoy, S.; Ay, R. Acaricide resistance and mechanisms in Tetranychus urticae populations from greenhouses in Turkey. Syst. Appl. Acarol. 2020, 25, 155–168. [Google Scholar]
- Bryon, A.; Kurlovs, A.H.; Dermauw, W.; Greenhalgh, R.; Riga, M.; Grbić, M.; Tirry, L.; Osakabe, M.; Vontas, J.; Clark, R.M.; et al. Disruption of a horizontally transferred phytoene desaturase abolishes carotenoid accumulation and diapause in Tetranychus urticae. Proc. Natl. Acad. Sci. USA 2017, 114, E5871–E5880. [Google Scholar] [CrossRef] [PubMed]
- Altincicek, B.; Kovacs, J.L.; Gerardo, N.M. Horizontally transferred fungal carotenoid genes in the two-spotted spider mite Tetranychus urticae. Biol. Lett. 2012, 8, 253–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toews, D.P.L.; Hofmeister, N.R.; Taylor, S.A. The evolution and genetics of carotenoid processing in animals. Trends Genet. 2017, 33, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Veerman, A. Carotenoid metabolism in Tetranychus urticae koch (Acari: Tetranychidae). Comp. Biochem. Physiol. B Comp. Biochem. 1974, 47, 101–116. [Google Scholar] [CrossRef]
- Metcalf, R.L.; Newell, I.M. Investigation of the biochromes of mites. Ann. Entomol. Soc. Am. 1962, 55, 350–353. [Google Scholar] [CrossRef]
- Wybouw, N.; Kurlovs, A.H.; Greenhalgh, R.; Bryon, A.; Kosterlitz, O.; Manabe, Y.; Osakabe, M.; Vontas, J.; Clark, R.M.; Van Leeuwen, T. Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191039. [Google Scholar] [CrossRef] [Green Version]
- Huo, S.M.; Zhang, Y.Y.; Song, Z.R.; Xiong, X.H.; Hong, X.Y. The potential pigmentation-related genes in spider mites revealed by comparative transcriptomes of the red form of Tetranychus urticae. Insect Mol. Biol. 2021, 30, 580–593. [Google Scholar] [CrossRef]
- Veerman, A. The pigments of Tetranychus cinnabarinus Boisd. (Acari: Tetranychidae). Comp. Biochem. Physiol. B Comp. Biochem. 1970, 36, 749–763. [Google Scholar] [CrossRef]
- Veerman, A. Carotenoids of wild-type and mutant strains of Tetranychus pacificus McGregor (Acari: Tetranychidae). Comp. Biochem. Physiol. B Comp. Biochem. 1972, 42, 329–340. [Google Scholar] [CrossRef]
- Bounfour, M.; Tanigoshi, L.K.; Chen, C.; Cameron, S.J.; Klauer, S. Chlorophyll content and chlorophyll fluorescence in red raspberry leaves infested with Tetranychus urticae and Eotetranychus carpini borealis (Acari: Tetranychidae). Environ. Entomol. 2002, 31, 215–220. [Google Scholar] [CrossRef]
- Iatrou, G.; Cook, C.M.; Stamou, G.; Lanaras, T. Chlorophyll fluorescence and leaf chlorophyll content of bean leaves injured by spider mites (Acari: Tetranychidae). Exp. Appl. Acarol. 1995, 19, 581–591. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Babani, F. Contents of photosynthetic pigments and ratios of chlorophyll a/b and chlorophylls to carotenoids (a plus b)/(x + c) in C4 plants as compared to C-3 plants. Photosynthetica 2022, 60, 3–9. [Google Scholar] [CrossRef]
- Mothes, U.; Seitz, K.A. Fine structure and function of the prosomal glands of the two-spotted spider mite, Tetranychus urticae (Acari, Tetranychidae). Cell Tissue Res. 1981, 221, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.J.; Andralojc, P.J.; Scales, J.C.; Salvucci, M.E.; Carmo-Silva, A.E.; Alonso, H.; Whitney, S.M. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2013, 64, 717–730. [Google Scholar] [CrossRef]
- Deans, C.A.; Behmer, S.T.; Fiene, J.; Sword, G.A. Spatio-temporal, genotypic, and environmental effects on plant soluble protein and digestible carbohydrate content: Implications for insect herbivores with cotton as an exemplar. J. Chem. Ecol. 2016, 42, 1151–1163. [Google Scholar] [CrossRef]
- Takaichi, S. General methods for identification of carotenoids. Biotechnol. Lett. 2014, 36, 1127–1128. [Google Scholar] [CrossRef]
- Krause, G.H.; Weis, E. Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 313–349. [Google Scholar] [CrossRef]
- Casazza, A.P.; Tarantino, D.; Soave, C. Preparation and functional characterization of thylakoids from Arabidopsis thaliana. Photosynth. Res. 2001, 68, 175–180. [Google Scholar] [CrossRef]
- Stirbet, A.; Govindjee. On the relation between the Kautsky effect (chlorophyll a fluorescence induction) and Photosystem II: Basics and applications of the OJIP fluorescence transient. J. Photochem. Photobiol. B Biol. 2011, 104, 236–257. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, F.; Béal, D.; Joliot, A.; Joliot, P. On the advantages of using green light to study fluorescence yield changes in leaves. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1767, 56–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailleul, B.; Cardol, P.; Breyton, C.; Finazzi, G. Electrochromism: A useful probe to study algal photosynthesis. Photosynth. Res. 2010, 106, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Sales, C.R.G.; Degen, G.E.; da Silva, A.B.; Carmo-Silva, E. Spectrophotometric determination of RuBisCO activity and activation state in leaf extracts. Meth. Mol. Biol. 2018, 1770, 239–250. [Google Scholar]











| Compound | RT (min) | Healthy Leaf | TSSM-Infested Leaf | TSSM |
|---|---|---|---|---|
| Chl b | 14.6 | 20.603 (0.619) a | 15.323 (0.511) b | 0.92 (0.034) c |
| Chl b’ | 15.1 | 1.298 (0.018) b | 0.661 (0.014) a | 0.622 (0.022) a |
| Chl a | 16.3 | 77.444 (1.472) a | 62.585 (2.447) b | 1.555 (0.048) c |
| Chl a’ | 16.9 | 15.185 (0.457) a | 9.789 (0.392) b | 1.963 (0.021) c |
| Chl a/b ratio | 3.75 | 4.08 | 1.68 | |
| Pheo b’ | 19.2 | n.d. | 1.005 (0.032) b | 5.809 (0.211) a |
| Pheo b | 19.5 | n.d. | 0.652 (0.024) b | 1.21 (0.017) a |
| Pheo a’ | 20.3 | 0.594 (0.009) b | 1.49 (0.016) b | 38.925 (1.188) a |
| Pheo a | 20.3 | 0.069 (0.002) a | 0.21 (0.004) b | 3.649 (0.083) c |
| TChP | 115.193 (2.577) a | 91.715 (3.44) b | 54.653 (1.624) c |
| Compound | RT (min) | Healthy Leaf | TSSM-Infested Leaf | TSSM |
|---|---|---|---|---|
| Lutein | 15.3 | 5.602 (0.014) a | 1.645 (0.034) b | n.d. |
| Putative xanthophyll | 18.6 | 5.154 (0.15) b | 5.464 (0.188) b | 44.545 (0.467) a |
| 15-cis-β-carotene | 20.8 | 2.822 (0.052) b | 2.108 (0.046) c | 22.588 (0.372) a |
| 13-cis-β-carotene | 21.5 | 8.646 (0.187) b | 6.671 (0.197) c | 48.061 (0.531) a |
| trans-α-carotene | 21.8 | 15.57 (0.167) b | 10.661 (0.263) c | 19.926 (0.502) a |
| cis-α-carotene | 22.6 | 1.192 (0.034) c | 8.006 (0.123) b | 9.723 (0.346) a |
| trans-β-carotene | 23.0 | 153.664 (3.95) b | 121.473 (4.561) c | 630.804 (24.929) a |
| 9-cis-β-carotene | 23.5 | 19.331 (0.632) a | 15.83 (0.364) b | n.d. |
| γ-carotene | 24.2 | 5.273 (0.019) b | 3.981 (0.253) c | 65.127 (0.595) a |
| TCrC | 222.453 (5.332) b | 179.286 (6.141) c | 849.482 (28.071) a |
| Polypeptide Size (kDa) | Healthy Leaves | TSSM-Infested Leaves | TSSM |
|---|---|---|---|
| 14.0 | 218.2 (±3.9 a) | 34.2 (±2.2 b) | n.c. |
| 18.0 | 29.2 (±1.2 a) | 10.0 (±2.9 b) | n.c. |
| 21.0 | 243.5 (±28.0 a) | 128.0 (±11.1 b) | 30.1 (±4.5 c) |
| 22.0 | 82.4 (±3.6) | n.c. | n.c. |
| 26.0 | 1224.5 (±144.0 a) | 747.9 (±32.0 b) | 179.9 (±14.4 c) |
| 37.0 | 51.9 (±5.7 a) | 262.7 (±13.3 b) | n.c. |
| 42.0 | 47.1 (±6.2 a) | n.c. | 8.3 (±2.9 b) |
| 46.0 | 4.1 (±1.0 a) | 6.8 (±3.0 a) | 63.5 (±5.4 b) |
| 56.0 | 364.5 (±21.2 a) | 179.6 (±11.6 b) | 3.6 (±0.5 c) |
| Total content | 2284.5 (±223.6 a) | 1319.0 (±117.1 b) | 279.2 (±18.0 c) |
| RubisCO | 582.7 (±24.1 a) | 202.4 (±18.8 b) | 3.6 (±0.5 c) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parmagnani, A.S.; Mannino, G.; Brillada, C.; Novero, M.; Dall’Osto, L.; Maffei, M.E. Biology of Two-Spotted Spider Mite (Tetranychus urticae): Ultrastructure, Photosynthesis, Guanine Transcriptomics, Carotenoids and Chlorophylls Metabolism, and Decoyinine as a Potential Acaricide. Int. J. Mol. Sci. 2023, 24, 1715. https://doi.org/10.3390/ijms24021715
Parmagnani AS, Mannino G, Brillada C, Novero M, Dall’Osto L, Maffei ME. Biology of Two-Spotted Spider Mite (Tetranychus urticae): Ultrastructure, Photosynthesis, Guanine Transcriptomics, Carotenoids and Chlorophylls Metabolism, and Decoyinine as a Potential Acaricide. International Journal of Molecular Sciences. 2023; 24(2):1715. https://doi.org/10.3390/ijms24021715
Chicago/Turabian StyleParmagnani, Ambra S., Giuseppe Mannino, Carla Brillada, Mara Novero, Luca Dall’Osto, and Massimo E. Maffei. 2023. "Biology of Two-Spotted Spider Mite (Tetranychus urticae): Ultrastructure, Photosynthesis, Guanine Transcriptomics, Carotenoids and Chlorophylls Metabolism, and Decoyinine as a Potential Acaricide" International Journal of Molecular Sciences 24, no. 2: 1715. https://doi.org/10.3390/ijms24021715
APA StyleParmagnani, A. S., Mannino, G., Brillada, C., Novero, M., Dall’Osto, L., & Maffei, M. E. (2023). Biology of Two-Spotted Spider Mite (Tetranychus urticae): Ultrastructure, Photosynthesis, Guanine Transcriptomics, Carotenoids and Chlorophylls Metabolism, and Decoyinine as a Potential Acaricide. International Journal of Molecular Sciences, 24(2), 1715. https://doi.org/10.3390/ijms24021715