Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression
Abstract
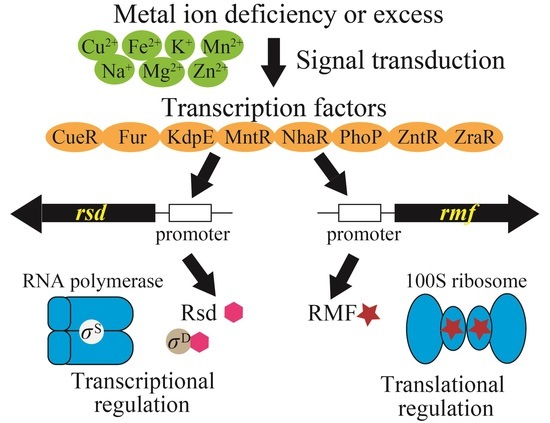
:1. Introduction
2. Results
2.1. Many Metal-Responsive TFs May Be Able to Bind to the Promoters of the rsd and rmf Genes
2.2. Genetic Defects in Some Metal-Responsive TFs Affect Rsd and Rmf Transcript Levels
2.3. Genetic Defects in Some Metal-Responsive TFs Affect Rsd and Rmf Protein Levels
2.4. Genetic Defects in Some Metal-Responsive TFs Affect 100S Ribosome Formation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Purification of TFs
4.3. Preparation of DNA Probes
4.4. PS-TF Screening System
4.5. Magnetic Bead Assays
4.6. Quantitative PCR (qPCR)
4.7. Western Blot Analysis
4.8. Measurement of 100S Ribosome Level Using Sucrose Density Gradient Centrifugation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishihama, A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000, 54, 499–518. [Google Scholar] [CrossRef]
- Jishage, M.; Ishihama, A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 1998, 95, 4953–4958. [Google Scholar] [CrossRef] [Green Version]
- Hengge-Aronis, R. Survival of hunger and stress; the role of rpoS in early stationary phase gene regulation in E. coli. Cell 1993, 72, 165–168. [Google Scholar] [CrossRef]
- Loewen, P.C.; Hengge-Aronis, R. The role of sigma factor σs (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 1994, 48, 53–80. [Google Scholar] [CrossRef]
- Wada, A.; Yamazaki, Y.; Fujita, N.; Ishihama, A. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc. Natl. Acad. Sci. USA 1990, 87, 2657–2661. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Wada, A. The 100S ribosome: Ribosomal hibernation induced by stress. Wiley Interdiscip. Rev. RNA 2014, 5, 723–732. [Google Scholar] [CrossRef]
- Yoshida, H.; Shimada, T.; Ishihama, A. Coordinated hibernation of transcriptional and translational apparatus during growth transition of Escherichia coli to stationary phase. mSystems 2018, 3, e00057-18. [Google Scholar] [CrossRef] [Green Version]
- Shimada, T.; Yoshida, H.; Ishihama, A. Involvement of cyclic AMP receptor protein in regulation of the rmf gene encoding the ribosome modulation factor in Escherichia coli. J. Bacteriol. 2013, 195, 2212–2219. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Wada, A.; Shimada, T.; Maki, Y.; Ishihama, A. Coordinated regulation of rsd and RMF for simultaneous hibernation of transcription apparatus and translation machinery in stationary-phase Escherichia coli. Front. Genet. 2019, 10, 1153. [Google Scholar] [CrossRef]
- van der Oost, J.; de Boer, A.P.; de Gier, J.W.; Zumft, W.G.; Stouthamer, A.H.; van Spanning, R.J. The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol. Lett. 1994, 121, 1–9. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Jenkinson, H.F. Out of the Iron Age: New insights into the critical role of manganese homeostasis in bacteria. Microbiol. Read. 2001, 147, 1709–1718. [Google Scholar] [CrossRef]
- Reeves, E.P.; Lu, H.; Jacobs, H.L.; Messina, C.G.; Bolsover, S.; Gabella, G.; Potma, E.O.; Warley, A.; Roes, J.; Segal, A.W. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 2002, 416, 291–297. [Google Scholar] [CrossRef]
- Carpenter, B.M.; Whitmire, J.M.; Merrell, D.S. This is not your mother’s repressor: The complex role of fur in pathogenesis. Infect. Immun. 2009, 77, 2590–2601. [Google Scholar] [CrossRef] [Green Version]
- Pruteanu, M.; Neher, S.B.; Baker, T.A. Ligand-controlled proteolysis of the Escherichia coli transcriptional regulator ZntR. J. Bacteriol. 2007, 189, 3017–3025. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.; Ishihama, A.; Busby, S.J.; Grainger, D.C. The Escherichia coli K-12 MntR miniregulon includes dps, which encodes the major stationary-phase DNA-binding protein. J. Bacteriol. 2011, 193, 1477–1480. [Google Scholar] [CrossRef] [Green Version]
- Bittner, L.M.; Kraus, A.; Schäkermann, S.; Narberhaus, F. The copper efflux regulator CueR is subject to ATP-dependent proteolysis in Escherichia coli. Front. Mol. Biosci. 2017, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Munson, G.P.; Lam, D.L.; Outten, F.W.; O’halloran, T.V. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 2000, 182, 5864–5871. [Google Scholar] [CrossRef] [Green Version]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef]
- Reyes-Caballero, H.; Campanello, G.C.; Giedroc, D.P. Metalloregulatory proteins: Metal selectivity and allosteric switching. Biophys. Chem. 2011, 156, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.; Norton, I.; Salunkhe, A.S.; Goodluck, H.; Aly, W.S.; Mourad-Agha, H.; Cornelis, P. Control of iron metabolism in bacteria. Met. Ions Life Sci. 2013, 12, 203–239. [Google Scholar] [CrossRef]
- Seo, S.W.; Kim, D.; Latif, H.; O’Brien, E.J.; Szubin, R.; Palsson, B.O. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014, 5, 4910. [Google Scholar] [CrossRef] [Green Version]
- Ogasawara, H.; Shinohara, S.; Yamamoto, K.; Ishihama, A. Novel regulation targets of the metal-response BasS-BasR two-component system of Escherichia coli. Microbiology (Reading) 2012, 158, 1482–1492. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Jin, J.; Liu, H.; Hao, Y.; Zhang, H.; Xie, Y. YbfA Regulates the sensitivity of Escherichia coli K12 to plantaricin BM-1 via the BasS/BasR two-component regulatory system. Front. Microbiol. 2021, 12, 659198. [Google Scholar] [CrossRef]
- Urano, H.; Yoshida, M.; Ogawa, A.; Yamamoto, K.; Ishihama, A.; Ogasawara, H. Cross-regulation between two common ancestral response regulators, HprR and CusR, in Escherichia coli. Microbiol. Read. 2017, 163, 243–252. [Google Scholar] [CrossRef]
- Outten, F.W.; Huffman, D.L.; Hale, J.A.; O’halloran, T.V. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 2001, 276, 30670–30677. [Google Scholar] [CrossRef] [Green Version]
- Wofford, J.D.; Bolaji, N.; Dziuba, N.; Outten, F.W.; Lindahl, P.A. Evidence that a respiratory shield in Escherichia coli protects a low-molecular-mass Fe(II) pool from O2-dependent oxidation. J. Biol. Chem. 2019, 294, 50–62. [Google Scholar] [CrossRef] [Green Version]
- Ballal, A.; Basu, B.; Apte, S.K. The Kdp-ATPase system and its regulation. J. Biosci. 2007, 32, 559–568. [Google Scholar] [CrossRef]
- Freeman, Z.N.; Dorus, S.; Waterfield, N.R. The KdpD/KdpE two-component system: Integrating K+ homeostasis and virulence. PLoS Pathog. 2013, 9, e1003201. [Google Scholar] [CrossRef] [Green Version]
- Anjem, A.; Varghese, S.; Imlay, J.A. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2009, 72, 844–858. [Google Scholar] [CrossRef] [Green Version]
- Król, J.E. Regulatory loop between the CsrA system and NhaR, a high salt/high pH regulator. PLoS ONE 2018, 13, e0209554. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Altman, R.; Eiteman, M.A.; Altman, E. Effect of overexpressing nhaA and nhaR on sodium tolerance and lactate production in Escherichia coli. J. Biol. Eng. 2013, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Minagawa, S.; Ogasawara, H.; Kato, A.; Yamamoto, K.; Eguchi, Y.; Oshima, T.; Mori, H.; Ishihama, A.; Utsumi, R. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 2003, 185, 3696–3702. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Li, Q.; Xue, M.; Wang, Z.; Tu, J.; Song, X.; Shao, Y.; Han, X.; Xue, T.; Liu, H.; et al. The role of the phoP transcriptional regulator on biofilm formation of avian pathogenic Escherichia coli. Avian Pathol. 2019, 48, 362–370. [Google Scholar] [CrossRef]
- Kim, H.; Jang, G.; Kim, B.G.; Yoon, Y. Modulation of the metal(loid) specificity of whole-cell bioreporters by genetic engineering of ZntR metal-binding loops. J. Microbiol. Biotechnol. 2020, 30, 681–688. [Google Scholar] [CrossRef]
- Sallai, L.; Tucker, P.A. Crystal structure of the central and C-terminal domain of the σ54-activator ZraR. J. Struct. Biol. 2005, 151, 160–170. [Google Scholar] [CrossRef]
- Shimada, T.; Ogasawara, H.; Kobayashi, I.; Kobayashi, N.; Ishihama, A. Single-target regulators constitute the minority group of transcription factors in Escherichia coli K-12. Front. Microbiol. 2021, 12, 697803. [Google Scholar] [CrossRef]
- Gilston, B.A.; Wang, S.; Marcus, M.D.; Canalizo-Hernández, M.A.; Swindell, E.P.; Xue, Y.; Mondragón, A.; O’halloran, T.V. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol. 2014, 12, e1001987. [Google Scholar] [CrossRef] [Green Version]
- Outten, C.E.; O’halloran, T.V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 2001, 292, 2488–2492. [Google Scholar] [CrossRef] [Green Version]
- RegulonDB. Available online: http://regulondb.ccg.unam.mx/ (accessed on 26 July 2021).
- Urano, H.; Umezawa, Y.; Yamamoto, K.; Ishihama, A.; Ogasawara, H. Cooperative regulation of the common target genes between H2O2-sensing YedVW and Cu2+-sensing CusSR in Escherichia coli. Microbiol. Read. 2015, 161, 729–738. [Google Scholar] [CrossRef]
- Shimada, T.; Ogasawara, H.; Ishihama, A. Single-target regulators form a minor group of transcription factors in Escherichia coli K-12. Nucleic Acids Res. 2018, 46, 3921–3936. [Google Scholar] [CrossRef] [Green Version]
- Paul, N.K.; Baksh, K.A.; Arias, J.F.; Zamble, D.B. The impact of a His-tag on DNA binding by RNA polymerase alpha-C-terminal domain from Helicobacter pylori. Protein Expr. Purif. 2020, 167, 105541. [Google Scholar] [CrossRef]
- Yoshida, H.; Maki, Y.; Furuike, S.; Sakai, A.; Ueta, M.; Wada, A. YqjD is an inner membrane protein associated with stationary-phase ribosomes in Escherichia coli. J. Bacteriol. 2012, 194, 4178–4183. [Google Scholar] [CrossRef] [Green Version]
- Ishihama, A. Prokaryotic genome regulation: Multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 2010, 34, 626–645. [Google Scholar] [CrossRef] [Green Version]
- Ogasawara, H.; Ishizuka, T.; Hotta, S.; Aoki, M.; Shimada, T.; Ishihama, A. Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12. Microbiol. Read. 2020, 166, 880–890. [Google Scholar] [CrossRef]
- Beauchene, N.A.; Mettert, E.L.; Moore, L.J.; Keles, S.; Willey, E.R.; Kiley, P.J. O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 12261–12266. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [Green Version]
- Vogel, H.J.; Bonner, D.M. Acetylornithinase of Escherichia coli; partial purification and some properties. J. Biol. Chem. 1956, 218, 97–106. [Google Scholar] [CrossRef]
- Shimada, K.; Ogasawara, H.; Yamada, K.; Shimura, M.; Kori, A.; Shimada, T.; Yamanaka, Y.; Yamamoto, K.; Ishihama, A. Screening of promoter-specific transcription factors: Multiple regulators for the sdiA gene involved in cell division control and quorum sensing. Microbiol. Read. 2013, 159, 2501–2512. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]






| TF | Mr(Da) | Family | Regulatory Function(s) | Effector | No. of Targets | Target TFs |
|---|---|---|---|---|---|---|
| BasR | 25,031 | OmpR | Biofilm formation and expression of outer membrane protein | TCS | 13–17 [22] | PutA, CsgD |
| CueR | 15,235 | MerR | Copper efflux regulator | Cu2+ | 2 [39] | - |
| CusR | 25,395 | OmpR | Efflux of copper and silver | TCS | 5 [40] | CusR, HprR |
| Fur | 16,795 | Fur | Ferric uptake regulator | Fe2+ | 132 [39] | FlhDC, Fur, MetJ, Nac, PurR, SoxS, SoxR |
| KdpE | 25,362 | OmpR | Potassium (K+) uptake | TCS | 1 [41] | KdpE |
| MntR | 17,640 | DtxR | Manganese transport regulator | Mn2+ | 5 [39] | Dps |
| NhaR | 34,284 | LysR | Adaptation to Na+ and alkaline pH, and biofilm formation | Na+ | 3 [39] | NhaR |
| PhoP | 25,535 | OmpR | Two-component regulatory system with PhoQ | TCS | 56 [39] | ArcA, GadE, GadW, PhoP, RstA, TreR |
| ZntR | 16,179 | MerR | Zn(II)-responsive regulator of zntA | Zn2+ | 1 [39] | - |
| ZraR | 48,395 | NtrC | Two-component regulatory system with ZraS | TCS | 3 [39] | ZraR |
| Zur | 19,254 | Fur | Zinc uptake regulator | Zn2+ | 6 [39] | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, H.; Shimada, T.; Ishihama, A. Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression. Int. J. Mol. Sci. 2023, 24, 4717. https://doi.org/10.3390/ijms24054717
Yoshida H, Shimada T, Ishihama A. Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression. International Journal of Molecular Sciences. 2023; 24(5):4717. https://doi.org/10.3390/ijms24054717
Chicago/Turabian StyleYoshida, Hideji, Tomohiro Shimada, and Akira Ishihama. 2023. "Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression" International Journal of Molecular Sciences 24, no. 5: 4717. https://doi.org/10.3390/ijms24054717
APA StyleYoshida, H., Shimada, T., & Ishihama, A. (2023). Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression. International Journal of Molecular Sciences, 24(5), 4717. https://doi.org/10.3390/ijms24054717