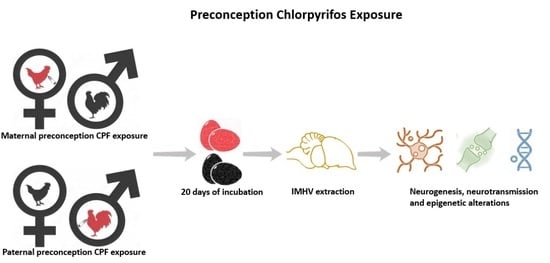
Parental Preconception and Pre-Hatch Exposure to a Developmental Insult Alters Offspring’s Gene Expression and Epigenetic Regulations: An Avian Model
Abstract
:1. Introduction
2. Results
2.1. Sex Determination
2.2. Paternal Exposure to Chlorpyrifos Alters Gene Expression in Offspring
2.3. Maternal Preconception Exposure to Chlorpyrifos Alters Gene Expression in the Offspring
2.3.1. Gene Expression Alterations
2.3.2. Chlorpyrifos Residues
2.4. Pre-Hatch Exposure to Chlorpyrifos Alters Gene Expression in the Offspring
2.5. Gene Co-Expression Correlation Network Analysis Reveals Disrupted Regulatory Network in Chlorpyrifos Affected Offspring
2.6. Egg and Body Weights of the Offspring
3. Discussion
4. Materials and Methods
4.1. Chicken Housing
4.2. Chlorpyrifos Administration
4.2.1. Pre-Hatch Exposure to Chlorpyrifos
4.2.2. Parental Preconception Exposure to Chlorpyrifos
4.3. Tissue Extraction
4.4. RNA Extraction
4.5. Real-Time qPCR Analysis
4.6. Sex Determination
4.7. Chlorpyrifos Residues
4.8. Egg and Body Weights of the Offspring
4.9. Statistical Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Werboff, J.; Gottlier, J.S. Drug in pregnancy: Behavioral teratology. Obstet. Gynecol. Surv. 1963, 18, 420–423. [Google Scholar] [CrossRef]
- Yanai, J.G.; Ginsburg, B.E. The effect of alcohol consumed by parent mice on the susceptibility to audiogenic seizure and the open-field behavior of their offspring. Behav. Genet. 1973, 3, 418. [Google Scholar]
- Izrael, M.; Van der Zee, E.A.; Slotkin, T.A.; Yanai, J. Cholinergic synaptic signaling mechanisms underlying behavioral teratogenicity: Effects of nicotine, chlorpyrifos, and heroin converge on protein kinase C translocation in the intermedial part of the hyperstriatum ventrale and on imprinting behavior in an avian model. J. Neurosci. Res. 2004, 78, 499–507. [Google Scholar]
- Yanai, J.; Sze, P.Y.; Ginsburg, B.E. Effects of aminergic drugs and glutamic acid on audiogenic seizures induced by early exposure to ethanol. Epilepsia 1975, 16, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Yanai, J.; Pick, C.G. Neuron transplantation reverses phenobarbital-induced behavioral birth defects in mice. Int. J. Dev. Neurosci. 1988, 6, 409–416. [Google Scholar] [CrossRef]
- Yanai, J.; Laxer, U.; Pick, C.G.; Trombka, D. Dopaminergic denervation reverses behavioral deficits induced by prenatal exposure to phenobarbital. Brain Res. Dev. Brain Res. 1989, 48, 255–261. [Google Scholar] [CrossRef]
- Yanai, J.; Vigoda, M.J.; Ornoy, A. Reversal of neurobehavioral teratogenicity in animal models and human: Three decades of progress. Brain Res. Bull. 2019, 150, 328–342. [Google Scholar] [CrossRef]
- Weller, C.V. The Blastophthoric Effect of Chronic Lead Poisoning: I. Introduction. Clinical and experimental observations from the Literature. J. Med. Res. 1915, 33, 271–293. [Google Scholar]
- Rimawi, I.; Ornoy, A.; Yanai, J. Paternal and/or maternal preconception-induced neurobehavioral teratogenicity in animal and human models. Brain Res. Bull. 2021, 174, 103–121. [Google Scholar] [CrossRef]
- Champroux, A.; Cocquet, J.; Henry-Berger, J.; Drevet, J.R.; Kocer, A. A Decade of Exploring the Mammalian Sperm Epigenome: Paternal Epigenetic and Transgenerational Inheritance. Front. Cell Dev. Biol 2018, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Billauer-Haimovitch, H.; Slotkin, T.A.; Dotan, S.; Langford, R.; Pinkas, A.; Yanai, J. Reversal of chlorpyrifos neurobehavioral teratogenicity in mice by nicotine administration and neural stem cell transplantation. Behav. Brain Res. 2009, 205, 499–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkey, A.B.; Holloway, Z.; Dean, C.; Koburov, R.; Slotkin, T.A.; Seidler, F.J.; Levin, E.D. Neurobehavioral anomalies in zebrafish after sequential exposures to DDT and chlorpyrifos in adulthood: Do multiple exposures interact? Neurotoxicol. Teratol. 2021, 87, 106985. [Google Scholar] [CrossRef] [PubMed]
- Thirugnanam, M.; Forgash, A.J. Environmental impact of mosquito pesticides: Toxicity and anticholinesterase activity of chlorpyrifos to fish in a salt marsh habitat. Arch. Env. Contam. Toxicol. 1977, 5, 415–425. [Google Scholar] [CrossRef]
- Koshlukova, S.E.R.; Pope, N.R. Chlorpyrifos. In Encyclopedia of Toxicology, 3rd ed.; Wexler, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 930–934. [Google Scholar]
- Buratti, F.M.; Volpe, M.T.; Meneguz, A.; Vittozzi, L.; Testai, E. CYP-specific bioactivation of four organophosphorothioate pesticides by human liver microsomes. Toxicol. Appl. Pharm. 2003, 186, 143–154. [Google Scholar] [CrossRef]
- Tang, J.; Cao, Y.; Rose, R.L.; Brimfield, A.A.; Dai, D.; Goldstein, J.A.; Hodgson, E. Metabolism of chlorpyrifos by human cytochrome P450 isoforms and human, mouse, and rat liver microsomes. Drug Metab. Dispos. 2001, 29, 1201–1204. [Google Scholar] [PubMed]
- FAO/WHO. Pesticide Residues in Food. In Proceedings of the Joint Meeting of the FAO Panel of Experts on Pesticides Residues in Food and the Environment and the WHO Core Assessment Group, Rome, Italy, 20–29 September 1999; Food and Agriculture Organization & World Health Organization: Rome, Italy, 1999; pp. 1–61. [Google Scholar]
- ATSDR. Interaction Profile for: Chlorpyrifos, Lead, Mercury, and Methylmercury; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 2006. [Google Scholar]
- Solomon, K.R.W.; Williams, W.M.; Mackay, D.; Purdy, J.; Giddings, J.M.; Giesy, J.P. Properties and Uses of Chlorpyrifos in the United States. In Reviews of Environmental Contamination and Toxicology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 231, pp. 13–34. [Google Scholar]
- Mahugija, J.C.; Chibura, P.E.; Lugwisha, E. Occurrence of organochlorine and organophosphorus pesticide residues in poultry feeds, raw and cooked eggs from selected farms in Ilala and Kibaha Districts, Tanzania. J. Appl. Sci. Environ. Manag. 2018, 22, 191–196. [Google Scholar] [CrossRef]
- Kobor, M.S.; Weinberg, J. Focus on: Epigenetics and fetal alcohol spectrum disorders. Alcohol Res. Health 2011, 34, 29–37. [Google Scholar]
- Ross, E.J.; Graham, D.L.; Money, K.M.; Stanwood, G.D. Developmental consequences of fetal exposure to drugs: What we know and what we still must learn. Neuropsychopharmacology 2015, 40, 61–87. [Google Scholar] [CrossRef] [Green Version]
- Turgeman, G.; Rimawi, I.; Heifetz, E.M.; Pinkas, A.; Pulver, D.; Altman, I.; Yanai, J. Reversal of prenatal heroin-induced alterations in hippocampal gene expression via transplantation of mesenchymal stem cells during adulthood. Neurotoxicol. Teratol. 2022, 90, 107063. [Google Scholar] [CrossRef]
- Chiu, K.C.; Sisca, F.; Ying, J.H.; Tsai, W.J.; Hsieh, W.S.; Chen, P.C.; Liu, C.Y. Prenatal chlorpyrifos exposure in association with PPARgamma H3K4me3 and DNA methylation levels and child development. Environ. Pollut. 2021, 274, 116511. [Google Scholar] [CrossRef]
- Ricceri, L.; Venerosi, A.; Capone, F.; Cometa, M.F.; Lorenzini, P.; Fortuna, S.; Calamandrei, G. Developmental neurotoxicity of organophosphorous pesticides: Fetal and neonatal exposure to chlorpyrifos alters sex-specific behaviors at adulthood in mice. Toxicol. Sci. 2006, 93, 105–113. [Google Scholar] [CrossRef]
- Stern, C.D. Gastrulation in the Chick. In Gastrulation: From Cells to Embryo; Stern, C.D., Ed.; Cold Spring Harbor Press: New York, NY, USA, 2004; pp. 219–232. [Google Scholar]
- Clayton, N.S.; Emery, N.J. Avian Models for Human Cognitive Neuroscience: A Proposal. Neuron 2015, 86, 1330–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deist, M.S.; Gallardo, R.A.; Bunn, D.A.; Dekkers, J.C.M.; Zhou, H.; Lamont, S.J. Resistant and susceptible chicken lines show distinctive responses to Newcastle disease virus infection in the lung transcriptome. BMC Genom. 2017, 18, 989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duguma, R.D.; Dana, N.; Yami, A. Marek’s disease vaccination opened the door to rear indigenous chickens of Ethiopia under confined management. Int. J. Appl. Res. Vet. Med. 2006, 4, 121–127. [Google Scholar]
- Pinkas, A.; Turgeman, G.; Tayeb, S.; Yanai, J. An avian model for ascertaining the mechanisms of organophosphate neuroteratogenicity and its therapy with mesenchymal stem cell transplantation. Neurotoxicol. Teratol. 2015, 50, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, F.; Sanchez-Amate, M.C.; Sanchez-Santed, F.; Cubero, I. Neuroanatomical targets of the organophosphate chlorpyrifos by c-fos immunolabeling. Toxicol. Sci. 2005, 84, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Dam, K.; Seidler, F.J.; Slotkin, T.A. Transcriptional biomarkers distinguish between vulnerable periods for developmental neurotoxicity of chlorpyrifos: Implications for toxicogenomics. Brain Res. Bull. 2003, 59, 261–265. [Google Scholar] [CrossRef]
- Ozdemir, S.; Altun, S.; Ozkaraca, M.; Ghosi, A.; Toraman, E.; Arslan, H. Cypermethrin, chlorpyrifos, deltamethrin, and imidacloprid exposure up-regulates the mRNA and protein levels of bdnf and c-fos in the brain of adult zebrafish (Danio rerio). Chemosphere 2018, 203, 318–326. [Google Scholar] [CrossRef]
- Song, X.; Seidler, F.J.; Saleh, J.L.; Zhang, J.; Padilla, S.; Slotkin, T.A. Cellular mechanisms for developmental toxicity of chlorpyrifos: Targeting the adenylyl cyclase signaling cascade. Toxicol. Appl. Pharm. 1997, 145, 158–174. [Google Scholar] [CrossRef]
- Gonzalez-Maciel, A.; Romero-Velazquez, R.M.; Alfaro-Rodriguez, A.; Sanchez Aparicio, P.; Reynoso-Robles, R. Prenatal exposure to oxcarbazepine increases hippocampal apoptosis in rat offspring. J. Chem. Neuroanat. 2020, 103, 101729. [Google Scholar] [CrossRef]
- Yanai, J.; Rosselli-Austin, L.; Tabakoff, B. Neuronal deficits in mice following prenatal exposure to phenobarbital. Exp. Neurol. 1979, 64, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Grecco, G.G.; Mork, B.E.; Huang, J.Y.; Metzger, C.E.; Haggerty, D.L.; Reeves, K.C.; Gao, Y.; Hoffman, H.; Katner, S.N.; Masters, A.R.; et al. Prenatal methadone exposure disrupts behavioral development and alters motor neuron intrinsic properties and local circuitry. eLife 2021, 10, e66230. [Google Scholar] [CrossRef] [PubMed]
- Altman, J. Experimental reorganization of the cerebellar cortes. V. Effects of early x-irradiation schedules that allow or prevent the acquisition of basket cells. J. Comp. Neurol. 1976, 165, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, T.A.; Skavicus, S.; Levin, E.D.; Seidler, F.J. Paternal Delta9-Tetrahydrocannabinol Exposure Prior to Mating Elicits Deficits in Cholinergic Synaptic Function in the Offspring. Toxicol. Sci. 2020, 174, 210–217. [Google Scholar] [CrossRef]
- Slotkin, T.A.; Stadler, A.; Skavicus, S.; Card, J.; Ruff, J.; Levin, E.D.; Seidler, F.J. Is There a Critical Period for the Developmental Neurotoxicity of Low-Level Tobacco Smoke Exposure? Toxicol. Sci. 2017, 155, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Ibn Lahmar Andaloussi, Z.; Taghzouti, K.; Abboussi, O. Behavioural and epigenetic effects of paternal exposure to cannabinoids during adolescence on offspring vulnerability to stress. Int. J. Dev. Neurosci. 2019, 72, 48–54. [Google Scholar] [CrossRef] [Green Version]
- Rompala, G.R.; Finegersh, A.; Slater, M.; Homanics, G.E. Paternal preconception alcohol exposure imparts intergenerational alcohol-related behaviors to male offspring on a pure C57BL/6J background. Alcohol 2017, 60, 169–177. [Google Scholar] [CrossRef] [Green Version]
- White, S.L.; Vassoler, F.M.; Schmidt, H.D.; Pierce, R.C.; Wimmer, M.E. Enhanced anxiety in the male offspring of sires that self-administered cocaine. Addict. Biol. 2015, 21, 802–810. [Google Scholar] [CrossRef] [Green Version]
- Zhai, L.; Li, Y.; Lan, X.; Ai, L. MicroRNA-10a-5p suppresses cancer proliferation and division in human cervical cancer by targeting BDNF. Exp. Med. 2017, 14, 6147–6151. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Li, H.; Mao, S.; Wang, H.; Zen, K.; Zhang, C.; Li, L. MicroRNA-29a modulates axon branching by targeting doublecortin in primary neurons. Protein Cell 2014, 5, 160–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanai, J.; Beer, A.; Huleihel, R.; Izrael, M.; Katz, S.; Levi, Y.; Rozenboim, I.; Yaniv, S.P.; Slotkin, T.A. Convergent effects on cell signaling mechanisms mediate the actions of different neurobehavioral teratogens: Alterations in cholinergic regulation of protein kinase C in chick and avian models. Ann. N. Y. Acad. Sci. 2004, 1025, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Zahalka, E.A.; Seidler, F.J.; Lappi, S.E.; McCook, E.C.; Yanai, J.; Slotkin, T.A. Deficits in development of central cholinergic pathways caused by fetal nicotine exposure: Differential effects on choline acetyltransferase activity and [3H]hemicholinium-3 binding. Neurotoxicol. Teratol. 1992, 14, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Steingart, R.A.; Silverman, W.F.; Barron, S.; Slotkin, T.A.; Awad, Y.; Yanai, J. Neural grafting reverses prenatal drug-induced alterations in hippocampal PKC and related behavioral deficits. Brain Res. Dev. Brain Res. 2000, 125, 9–19. [Google Scholar] [CrossRef]
- Ben-Shaanan, T.L.; Ben-Hur, T.; Yanai, J. Transplantation of neural progenitors enhances production of endogenous cells in the impaired brain. Mol. Psychiatry 2008, 13, 222–231. [Google Scholar] [CrossRef] [Green Version]
- Turgeman, G.; Pinkas, A.; Slotkin, T.; Tfillin, M.; Langford, R.; Yanai, J. Reversal of chlorpyrifos neurobehavioral teratogenicity in mice by allographic transplantation of adult subventricular zone-derived neural stem cells. J. Neurosci. Res. 2011, 89, 1185–1193. [Google Scholar] [CrossRef]
- Miranda, M.; Morici, J.F.; Zanoni, M.B.; Bekinschtein, P. Brain-Derived Neurotrophic Factor: A Key Molecule for Memory in the Healthy and the Pathological Brain. Front. Cell Neurosci. 2019, 13, 363. [Google Scholar] [CrossRef]
- Rao, M.S.; Shetty, A.K. Efficacy of doublecortin as a marker to analyse the absolute number and dendritic growth of newly generated neurons in the adult dentate gyrus. Eur. J. Neurosci. 2004, 19, 234–246. [Google Scholar] [CrossRef]
- Brown, J.P.; Couillard-Despres, S.; Cooper-Kuhn, C.M.; Winkler, J.; Aigner, L.; Kuhn, H.G. Transient expression of doublecortin during adult neurogenesis. J. Comp. Neurol. 2003, 467, 1–10. [Google Scholar] [CrossRef]
- Couillard-Despres, S.; Winner, B.; Schaubeck, S.; Aigner, R.; Vroemen, M.; Weidner, N.; Bogdahn, U.; Winkler, J.; Kuhn, H.G.; Aigner, L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005, 21, 1–14. [Google Scholar] [CrossRef]
- Routtenberg, A. Protein kinase C activation leading to protein F1 phosphorylation may regulate synaptic plasticity by presynaptic terminal growth. Behav. Neural. Biol. 1985, 44, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.J.; Ji, R.R. c-Fos and pERK, which is a better marker for neuronal activation and central sensitization after noxious stimulation and tissue injury? Open Pain J. 2009, 2, 11–17. [Google Scholar] [CrossRef]
- O’Neill, C.E.; Newsom, R.J.; Stafford, J.; Scott, T.; Archuleta, S.; Levis, S.C.; Spencer, R.L.; Campeau, S.; Bachtell, R.K. Adolescent caffeine consumption increases adulthood anxiety-related behavior and modifies neuroendocrine signaling. Psychoneuroendocrinology 2016, 67, 40–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llado-Pelfort, L.; Santana, N.; Ghisi, V.; Artigas, F.; Celada, P. 5-HT1A receptor agonists enhance pyramidal cell firing in prefrontal cortex through a preferential action on GABA interneurons. Cereb. Cortex 2012, 22, 1487–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macchione, A.F.; Trujillo, V.; Anunziata, F.; Sahonero, M.; Anastasia, A.; Abate, P.; Molina, J.C. Early ethanol pre-exposure alters breathing patterns by disruptions in the central respiratory network and serotonergic balance in neonate rats. Behav. Brain Res. 2021, 396, 112908. [Google Scholar] [CrossRef]
- Ramamoorthy, S.; Bauman, A.L.; Moore, K.R.; Han, H.; Yang-Feng, T.; Chang, A.S.; Ganapathy, V.; Blakely, R.D. Antidepressant- and cocaine-sensitive human serotonin transporter: Molecular cloning, expression, and chromosomal localization. Proc. Natl. Acad. Sci. USA 1993, 90, 2542–2546. [Google Scholar] [CrossRef] [Green Version]
- Lam, D.; Ancelin, M.L.; Ritchie, K.; Freak-Poli, R.; Saffery, R.; Ryan, J. Genotype-dependent associations between serotonin transporter gene (SLC6A4) DNA methylation and late-life depression. BMC Psychiatry 2018, 18, 282. [Google Scholar] [CrossRef] [Green Version]
- Arvidsson, U.; Riedl, M.; Elde, R.; Meister, B. Vesicular acetylcholine transporter (VAChT) protein: A novel and unique marker for cholinergic neurons in the central and peripheral nervous systems. J. Comp. Neurol. 1997, 378, 454–467. [Google Scholar] [CrossRef]
- Van der Zee, E.A.; Bolhuis, J.J.; Solomonia, R.O.; Horn, G.; Luiten, P.G. Differential distribution of protein kinase C (PKC alpha beta and PKC gamma) isoenzyme immunoreactivity in the chick brain. Brain Res. 1995, 676, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Errico, M.C.; Felicetti, F.; Bottero, L.; Mattia, G.; Boe, A.; Felli, N.; Petrini, M.; Bellenghi, M.; Pandha, H.S.; Calvaruso, M.; et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int. J. Cancer 2013, 133, 879–892. [Google Scholar] [CrossRef] [Green Version]
- Godschalk, R.; Remels, A.; Hoogendoorn, C.; van Benthem, J.; Luijten, M.; Duale, N.; Brunborg, G.; Olsen, A.K.; Bouwman, F.G.; Munnia, A.; et al. Paternal Exposure to Environmental Chemical Stress Affects Male Offspring's Hepatic Mitochondria. Toxicol. Sci. 2018, 162, 241–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Sun, D.; Jakovcevski, M.; Jiang, Y. Epigenetic mechanism of SETDB1 in brain: Implications for neuropsychiatric disorders. Transl. Psychiatry 2020, 10, 115. [Google Scholar] [CrossRef] [Green Version]
- Wood, K.H.; Zhou, Z. Emerging Molecular and Biological Functions of MBD2, a Reader of DNA Methylation. Front. Genet. 2016, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Schoenherr, C.J.; Anderson, D.J. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 1995, 267, 1360–1363. [Google Scholar] [CrossRef]
- Chong, J.A.; Tapia-Ramirez, J.; Kim, S.; Toledo-Aral, J.J.; Zheng, Y.; Boutros, M.C.; Altshuller, Y.M.; Frohman, M.A.; Kraner, S.D.; Mandel, G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995, 80, 949–957. [Google Scholar] [CrossRef] [Green Version]
- Buffolo, F.; Petrosino, V.; Albini, M.; Moschetta, M.; Carlini, F.; Floss, T.; Kerlero de Rosbo, N.; Cesca, F.; Rocchi, A.; Uccelli, A.; et al. Neuroinflammation induces synaptic scaling through IL-1beta-mediated activation of the transcriptional repressor REST/NRSF. Cell Death Dis. 2021, 12, 180. [Google Scholar] [CrossRef]
- Bourtchuladze, R.; Frenguelli, B.; Blendy, J.; Cioffi, D.; Schutz, G.; Silva, A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 1994, 79, 59–68. [Google Scholar] [CrossRef]
- Kitagawa, H.; Sugo, N.; Morimatsu, M.; Arai, Y.; Yanagida, T.; Yamamoto, N. Activity-Dependent Dynamics of the Transcription Factor of cAMP-Response Element Binding Protein in Cortical Neurons Revealed by Single-Molecule Imaging. J. Neurosci. 2017, 37, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Landeira, B.S.; Santana, T.; Araujo, J.A.M.; Tabet, E.I.; Tannous, B.A.; Schroeder, T.; Costa, M.R. Activity-Independent Effects of CREB on Neuronal Survival and Differentiation during Mouse Cerebral Cortex Development. Cereb. Cortex 2018, 28, 538–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberini, C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.A.; Krawetz, S.A. RNA in spermatozoa: Implications for the alternative haploid genome. Mol. Hum. Reprod. 1997, 3, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Krawetz, S.A.; Kruger, A.; Lalancette, C.; Tagett, R.; Anton, E.; Draghici, S.; Diamond, M.P. A survey of small RNAs in human sperm. Hum. Reprod. 2011, 26, 3401–3412. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.; Ostermeier, G.C.; Krawetz, S.A. The controversy, potential and roles of spermatozoal RNA. Trends Mol. Med. 2005, 11, 156–163. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Miller, D.; Huntriss, J.D.; Diamond, M.P.; Krawetz, S.A. Reproductive biology: Delivering spermatozoan RNA to the oocyte. Nature 2004, 429, 154. [Google Scholar] [CrossRef]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef] [Green Version]
- Wykes, S.M.; Miller, D.; Krawetz, S.A. Mammalian spermatozoal mRNAs: Tools for the functional analysis of male gametes. J. Submicrosc. Cytol. Pathol. 2000, 32, 77–81. [Google Scholar]
- Wykes, S.M.; Visscher, D.W.; Krawetz, S.A. Haploid transcripts persist in mature human spermatozoa. Mol. Hum. Reprod. 1997, 3, 15–19. [Google Scholar] [CrossRef]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016, 143, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Reilly, J.N.; McLaughlin, E.A.; Stanger, S.J.; Anderson, A.L.; Hutcheon, K.; Church, K.; Mihalas, B.P.; Tyagi, S.; Holt, J.E.; Eamens, A.L.; et al. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci. Rep. 2016, 6, 31794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutcheon, K.; McLaughlin, E.A.; Stanger, S.J.; Bernstein, I.R.; Dun, M.D.; Eamens, A.L.; Nixon, B. Analysis of the small non-protein-coding RNA profile of mouse spermatozoa reveals specific enrichment of piRNAs within mature spermatozoa. RNA Biol. 2017, 14, 1776–1790. [Google Scholar] [CrossRef] [PubMed]
- Pantano, L.; Jodar, M.; Bak, M.; Ballesca, J.L.; Tommerup, N.; Oliva, R.; Vavouri, T. The small RNA content of human sperm reveals pseudogene-derived piRNAs complementary to protein-coding genes. RNA 2015, 21, 1085–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mychasiuk, R.; Harker, A.; Ilnytskyy, S.; Gibb, R. Paternal stress prior to conception alters DNA methylation and behaviour of developing rat offspring. Neuroscience 2013, 241, 100–105. [Google Scholar] [CrossRef]
- De Reviers, M. De´Termination de la Dure´e des Processus Spermatoge´Netiques Chez le coq a` L’aide de Thymidine Tritie´e. In Proceedings of the Congres International de Reproduction Animale et Insemination Artificielle Paris 1, Paris, France, 22–26 July 1968; pp. 183–185. [Google Scholar]
- Marchand, C.R.; Gomot, L.; de Reviers, M. Etude par autoradiographie et Marquage a` la thymidine tritie´e de la dure´e de la spermatoge´ne`se du canard de barbarie (Cairina moschata L.); Comptes Rendus des Se´ances de la Socie´te´ de Biologie et de ses Filiales: Paris, France, 1977; pp. 931–934. [Google Scholar]
- Blesbois, E.; Brillard, J.P. Specific features of in vivo and in vitro sperm storage in birds. Animal 2007, 1, 1472–1481. [Google Scholar] [CrossRef] [Green Version]
- Noirault, J.; Brillard, J.P.; Bakst, M.R. Spermatogenesis in the turkey (Meleagris gallopavo): Quantitative approach in immature and adult males subjected to various photoperiods. Theriogenology 2006, 65, 845–859. [Google Scholar] [CrossRef] [Green Version]
- Rzymski, P.; Rzymski, P.; Tomczyk, K.; Niedzielski, P.; Jakubowski, K.; Poniedzialek, B.; Opala, T. Metal status in human endometrium: Relation to cigarette smoking and histological lesions. Env. Res. 2014, 132, 328–333. [Google Scholar] [CrossRef]
- Nolan, R.J.; Rick, D.L.; Freshour, N.L.; Saunders, J.H. Chlorpyrifos: Pharmacokinetics in human volunteers. Toxicol. Appl. Pharm. 1984, 73, 8–15. [Google Scholar] [CrossRef]
- Shah, P.V.; Monroe, R.J.; Guthrie, F.E. Comparative rates of dermal penetration of insecticides in mice. Toxicol. Appl. Pharm. 1981, 59, 414–423. [Google Scholar] [CrossRef]
- Dubiec, A.; Zagalska-Neubauer, M. Molecular techniques for sex identification in birds. Biol. Lett. 2006, 43, 3–12. [Google Scholar]
- Wan, Z.; Lu, Y.; Rui, L.; Yu, X.; Li, Z. Sexing chick mRNA: A protocol based on quantitative real-time polymerase chain reaction. Poult. Sci. 2017, 96, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, N.F.; Ellis, H.; Shioda, K.; Mahoney, C.; Coser, K.R.; Shioda, T. High-throughput applicable genomic sex typing of chicken by TaqMan real-time quantitative polymerase chain reaction. Poult. Sci. 2010, 89, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.; Double, M.C.; Orr, K.; Dawson, R.J. A DNA test to sex most birds. Mol. Ecol. 1998, 7, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Horn, G. Imprinting, learning, and memory. Behav. Neurosci. 1986, 100, 825–832. [Google Scholar] [CrossRef]
- McLennan, J.G.; Horn, G. Learning-dependent Changes in the Responses to Visual Stimuli of Neurons in a Recognition Memory System. Eur. J. Neurosci. 1992, 4, 1112–1122. [Google Scholar] [CrossRef]
- National Research Council. Guide for the Care and Use of Laboratory Animals, 8th ed.; The National Academies Collection: Reports funded by National Institutes of Health; The National Accademise Press: Washington, DC, USA, 2011. [Google Scholar]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef]
- Pearson, R.T. The Avian Brain; Academic Press: New York, NY, USA, 1972. [Google Scholar]
- Reiner, A.; Perkel, D.J.; Bruce, L.L.; Butler, A.B.; Csillag, A.; Kuenzel, W.; Medina, L.; Paxinos, G.; Shimizu, T.; Striedter, G.; et al. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 2004, 473, 377–414. [Google Scholar] [CrossRef]
- Horn, G. Technique for removing IMHV from the chick brain. In Neural and Behavioural Plasticity. The Use of the Domestic Chick as a Model; Andrew, R., Ed.; Oxford University Press: Oxford, UK, 1991; Volume 1, pp. 44–48. [Google Scholar]
- Tfilin, M.; Merenlender, A.; Gispan, I.; Yadid, G.; Turgeman, G. Mesenchymal Stem Cells Increase Hippocampal Neurogenesis and Counteract Depressive-Like Behavior. Mol. Psychiatry 2010, 15, 1164–1175. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Britzi, M.; Perucca, E.; Soback, S.; Levy, R.H.; Fattore, C.; Crema, F.; Gatti, G.; Doose, D.R.; Maryanoff, B.E.; Bialer, M. Pharmacokinetic and metabolic investigation of topiramate disposition in healthy subjects in the absence and in the presence of enzyme induction by carbamazepine. Epilepsia 2005, 46, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]










| Exposure | Treatment | #Egg Weight (n) | Body Weight (n) |
|---|---|---|---|
| Pre-hatch | Control | 43.6 ± 0.6 (45) | 29.5 ± 1.3 (27) |
| Pre-hatch | CPF | 43.9 ± 0.6 (45) | 29.8 ± 1.5 (21) |
| Maternal | Control | 35.5 ± 0.9 (45) | 20.1 ± 0.3 (46) |
| Maternal | CPF | 39.8 ± 0.8 * (53) | 21.2 ± 0.3 * (53) |
| Paternal | Control | 43.8 ± 0.6 (46) | 26.2 ± 0.7 (47) |
| Paternal | CPF | 44.2 ± 0.4 (41) | 24.4 ± 0.6 (39) |
| Primer | Sequence (5′-3′) |
|---|---|
| DCX Forward | AGAAGACGGCCCATTCGTTT |
| DCX Reverse | GGTCACCTGCTTTCCATCCATCCA |
| PKCß Forward | CAAACGCCATTTCTAAGTTCGA |
| PKCß Reverse | CAGCATCACCTTCCCAAAGC |
| BDNF Forward | AGCAGTCAAGTGCCTTTGGAA |
| BDNF Reverse | AGTGACGCCGGACTCTCATG |
| FOS Forward | ACCTCCTCCAGAGATGTAG |
| FOS Reverse | AGCACCAGTTAATTCCAATC |
| CHRM2 Forward | TGGAACATAACAAAGTCCAG |
| CHRM2 Reverse | TTTGTATTGGAAGGAACCAC |
| CHRM3 Forward | TGGTATATCCAACTACAGGC |
| CHRM3 Reverse | ACAACTGGTAGTGTGTCAG |
| SLC18A3 Forward | GATAGACCCCTATATCGCC |
| SLC18A3 Reverse | CATGGACTCCTTCATCCAG |
| SLC6A4 Forward | ATAATGCATGGAACACAGG |
| SLC6A4 Reverse | TGGCGGGTATAAAATTCTTC |
| CHDZ Forward | AGTCAGGCAGTCAATCCGAA |
| CHDZ Reverse | CCACTGTCTGATGATGCTGC |
| CHDW Forward | GGGATTCTGAGTGAAGACTGG |
| CHDW Reverse | CTGTCCTGTGCCCTTCTTG |
| MBD2 Forward | CATCCCAATAAGGGCAAAC |
| MBD2 Reverse | GGGTTGCTTAAAGATGGAC |
| MBD3 Forward | AAGAAGGAGAAGATGAGGAG |
| MBD3 Reverse | GAATTCTTAGATGGAACTCCC |
| MeCP2 Forward | GGACCAGGAAGCTCAAACAG |
| MeCP2 Reverse | GAAGTACGCGATCAGTTCCAC |
| SETDB1 Forward | CTCCTTCGTCTGCATCTACG |
| SETDB1 Reverse | AAGTATTCGTCGCCCATCTC |
| SETDB2 Forward | AGACAGGCAAAACAAGGCATA |
| SETDB2 Reverse | AGCAGCTGTGATTAAGAAAACG |
| REST Forward | AGAGGAAACCAGTCCGAAGA |
| REST Reverse | CATTGACCAAGTGGCGATT |
| CREB Forward | GAGAGAATAAAACTGCGGC |
| CREB Reverse | CCATGGTCATTTAGTTACCG |
| GAPDH Forward | TGGAGCCCCTGCTCTTCA |
| GAPDH Reverse | GGAACAGAACTGGCCTCTCACT |
| miR-221 Forward | AGCTACATTGTCTGCTGGGTTTC |
| miR-29a Forward | TAGCACCATTTGAAATCGGTT |
| miR-6612 Forward | ATGCTGTGTGTGCGTTCGTA |
| miR-10a Forward | TGTGTAAAGGAAGTTGGGTCACA |
| RNU6 Forward | GCAAATTCGTGAAGCGTTCC |
| Universal microRNA Reverse Primer | GCATAGACCTGAATGGCGGTA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rimawi, I.; Turgeman, G.; Avital-Cohen, N.; Rozenboim, I.; Yanai, J. Parental Preconception and Pre-Hatch Exposure to a Developmental Insult Alters Offspring’s Gene Expression and Epigenetic Regulations: An Avian Model. Int. J. Mol. Sci. 2023, 24, 5047. https://doi.org/10.3390/ijms24055047
Rimawi I, Turgeman G, Avital-Cohen N, Rozenboim I, Yanai J. Parental Preconception and Pre-Hatch Exposure to a Developmental Insult Alters Offspring’s Gene Expression and Epigenetic Regulations: An Avian Model. International Journal of Molecular Sciences. 2023; 24(5):5047. https://doi.org/10.3390/ijms24055047
Chicago/Turabian StyleRimawi, Issam, Gadi Turgeman, Nataly Avital-Cohen, Israel Rozenboim, and Joseph Yanai. 2023. "Parental Preconception and Pre-Hatch Exposure to a Developmental Insult Alters Offspring’s Gene Expression and Epigenetic Regulations: An Avian Model" International Journal of Molecular Sciences 24, no. 5: 5047. https://doi.org/10.3390/ijms24055047
APA StyleRimawi, I., Turgeman, G., Avital-Cohen, N., Rozenboim, I., & Yanai, J. (2023). Parental Preconception and Pre-Hatch Exposure to a Developmental Insult Alters Offspring’s Gene Expression and Epigenetic Regulations: An Avian Model. International Journal of Molecular Sciences, 24(5), 5047. https://doi.org/10.3390/ijms24055047