Metabolomic and Proteomic Profiling of Porcine Intestinal Epithelial Cells Infected with Porcine Epidemic Diarrhea Virus
Abstract
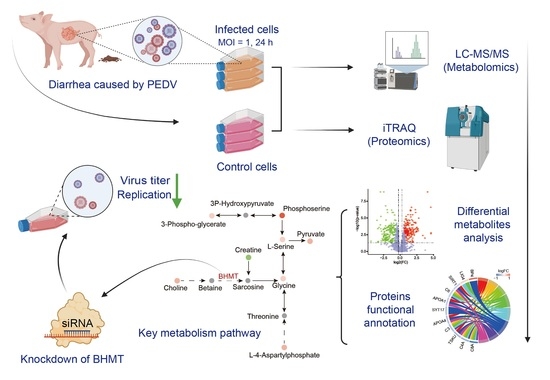
:1. Introduction
2. Results
2.1. Metabolomic Changes Associated with PEDV Infection
2.2. Alterations in Proteome Induced by PEDV Infection
2.3. Integrated Analysis of Metabolome and Proteome Data
2.4. Knockdown of BHMT Reduces PEDV Replication in Host Cells
3. Discussion
4. Materials and Methods
4.1. Cell Sample Preparation
4.2. Metabolite Identification by LC–MS/MS
4.3. Proteomic Analysis by iTRAQ
4.4. Integrated Analysis of Metabolome and Proteome
4.5. Functional Annotation for Differentially Expressed Proteins and Differential Metabolites
4.6. Knockdown of BHMT Expression by siRNA
4.7. qRT-PCR
4.8. Titration of PEDV
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niederwerder, M.C.; Hesse, R.A. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018, 65, 660–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrara, G.; D’Anza, E.; Rossi, A.; Improda, E.; Iovane, V.; Pagnini, U.; Iovane, G.; Montagnaro, S. A Serological Investigation of Porcine Reproductive and Respiratory Syndrome and Three Coronaviruses in the Campania Region, Southern Italy. Viruses 2023, 15, 300. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286, 198045. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Huang, L.; Yuan, C.; Wang, J.; Yang, Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018, 9, 3811. [Google Scholar] [CrossRef] [Green Version]
- Carvajal, A.; Argüello, H.; Martínez-Lobo, F.J.; Costillas, S.; Miranda, R.; de Nova, P.J.; Rubio, P. Porcine epidemic diarrhea: New insights into an old disease. Porc. Health Manag. 2015, 1, 12. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhu, Z.; Yang, F.; Cao, W.; Yang, J.; Ma, C.; Zhao, Z.; Tian, H.; Liu, X.; Ma, J.; et al. Porcine Epidemic Diarrhea Virus Membrane Protein Interacted with IRF7 to Inhibit Type I IFN Production during Viral Infection. J. Immunol. 2021, 206, 2909–2923. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Peng, Q.; Song, S.; Shi, D.; Zhang, X.; Guo, W.; Li, Y.; Zhou, J.; Zhu, X.; Zhao, Y.; et al. Nonstructural Protein 1 of Variant PEDV Plays a Key Role in Escaping Replication Restriction by Complement C3. J. Virol. 2022, 96, e0102422. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Thaker, S.K.; Ch’ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef]
- Zhou, L.; He, R.; Fang, P.; Li, M.; Yu, H.; Wang, Q.; Yu, Y.; Wang, F.; Zhang, Y.; Chen, A.; et al. Hepatitis B virus rigs the cellular metabolome to avoid innate immune recognition. Nat. Commun. 2021, 12, 98. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Song, Y.; Li, Z.; Xue, Z.; Cao, W.; Liu, X.; Zheng, H. African Swine Fever Virus Regulates Host Energy and Amino Acid Metabolism to Promote Viral Replication. J. Virol. 2022, 96, e0191921. [Google Scholar] [CrossRef]
- Garrow, T.A. Purification, kinetic properties, and cDNA cloning of mammalian betaine-homocysteine methyltransferase. J. Biol. Chem. 1996, 271, 22831–22838. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Wang, Q.; Ghneim, K.; Wang, L.; Rampanelli, E.; Holley-Guthrie, E.; Cheng, L.; Garrido, C.; Margolis, D.M.; Eller, L.A.; et al. Multi-omics analyses reveal that HIV-1 alters CD4+ T cell immunometabolism to fuel virus replication. Nat. Immunol. 2021, 22, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Liu, W.; Ding, X.; Liang, S.; Zheng, Y.; Zhu, X.; Quan, S.; Yi, X.; Xiang, N.; Du, J.; et al. Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19. Cell Rep. 2022, 38, 110271. [Google Scholar] [CrossRef]
- Nunes, A.; Ribeiro, D.R.; Marques, M.; Santos, M.A.S.; Ribeiro, D.; Soares, A.R. Emerging Roles of tRNAs in RNA Virus Infections. Trends Biochem. Sci. 2020, 45, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Passalacqua, K.D.; Lu, J.; Goodfellow, I.; Kolawole, A.O.; Arche, J.R.; Maddox, R.J.; Carnahan, K.E.; O’Riordan, M.X.D.; Wobus, C.E. Glycolysis Is an Intrinsic Factor for Optimal Replication of a Norovirus. mBio 2019, 10, e02175-18. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.Y.; Yang, Y.L.; Tang, Y.X.; Qin, P.; Wang, G.; Xie, J.Y.; Chen, S.X.; Ding, C.; Huang, Y.W.; Zhu, S.J. Bile acids promote the caveolae-associated entry of swine acute diarrhea syndrome coronavirus in porcine intestinal enteroids. PLoS Pathog. 2022, 18, e1010620. [Google Scholar] [CrossRef]
- Pearce, S.C.; Schweer, W.P.; Schwartz, K.J.; Yoon, K.J.; Lonergan, S.M.; Gabler, N.K. Pig jejunum protein profile changes in response to a porcine epidemic diarrhea virus challenge. J. Anim. Sci. 2016, 94, 412–415. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Chen, F.; Ye, S.; Guo, X.; Muhanmmad Memon, A.; Wu, M.; He, Q. Comparative Proteome Analysis of Porcine Jejunum Tissues in Response to a Virulent Strain of Porcine Epidemic Diarrhea Virus and Its Attenuated Strain. Viruses 2016, 8, 323. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Li, B.; Chen, L.; Ma, Z.; He, K.; Fan, H. Differential Protein Analysis of IPEC-J2 Cells Infected with Porcine Epidemic Diarrhea Virus Pandemic and Classical Strains Elucidates the Pathogenesis of Infection. J. Proteome Res. 2016, 16, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Lv, X.; Thompson, E.W.; Ostrikov, K.K. Histone lactylation: Epigenetic mark of glycolytic switch. Trends Genet. 2022, 38, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, P.J.; Davies, A. Complement and immunity to viruses. Immunol. Rev. 1997, 159, 69–77. [Google Scholar] [CrossRef]
- Kwon, Y.C.; Ray, R. Complement Regulation and Immune Evasion by Hepatitis C Virus. Methods Mol. Biol. 2019, 1911, 337–347. [Google Scholar]
- Banerjee, A.; Mazumdar, B.; Meyer, K.; Di Bisceglie, A.M.; Ray, R.B.; Ray, R. Transcriptional repression of C4 complement by hepatitis C virus proteins. J. Virol. 2011, 85, 4157–4166. [Google Scholar] [CrossRef] [Green Version]
- Avirutnan, P.; Fuchs, A.; Hauhart, R.E.; Somnuke, P.; Youn, S.; Diamond, M.S.; Atkinson, J.P. Inhibition of the human complement component c4 by flavivirus non-structural protein ns1. Mol. Immunol. 2010, 47, 2239. [Google Scholar] [CrossRef]
- Xie, C.B.; Jane-Wit, D.; Pober, J.S. Complement Membrane Attack Complex: New Roles, Mechanisms of Action, and Therapeutic Targets. Am. J. Pathol. 2020, 190, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.N.; da Silva, E.M.; Allonso, D.; Coelho, D.R.; Andrade, I.D.S.; de Medeiros, L.N.; Menezes, J.L.; Barbosa, A.S.; Mohana-Borges, R. Inhibition of the Membrane Attack Complex by Dengue Virus NS1 through Interaction with Vitronectin and Terminal Complement Proteins. J. Virol. 2016, 90, 9570–9581. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.H.; Gao, J.; Liu, X.D.; Tang, H.W.; Cao, S.L.; Zhang, J.K.; Wen, P.H.; Wang, Z.H.; Li, J.; Guo, W.Z.; et al. Interaction analysis of gene variants related to one-carbon metabolism with chronic hepatitis B infection in Chinese patients. J. Gene Med. 2021, 23, e3347. [Google Scholar] [CrossRef]
- Avila, M.A.; Berasain, C.; Torres, L.; Martín-Duce, A.; Corrales, F.J.; Yang, H.; Prieto, J.; Lu, S.C.; Caballería, J.; Rodés, J.; et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 2000, 3, 907–914. [Google Scholar] [CrossRef] [Green Version]
- Ou, X.; Yang, H.; Ramani, K.; Ara, A.I.; Chen, H.; Mato, J.M.; Lu, S.C. Inhibition of human betaine-homocysteine methyltransferase expression by S-adenosylmethionine and methylthioadenosine. Biochem. J. 2007, 401, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, Y.; Zhou, J.; Wang, X.; Ma, L.; Li, J.; Yang, L.; Yuan, H.; Pang, D.; Ouyang, H. Porcine Epidemic Diarrhea Virus: An Updated Overview of Virus Epidemiology, Virulence Variation Patterns and Virus-Host Interactions. Viruses 2022, 14, 2434. [Google Scholar] [CrossRef] [PubMed]
- Sellick, C.A.; Hansen, R.; Stephens, G.M.; Goodacre, R.; Dickson, A.J. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat. Protoc. 2011, 6, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef] [Green Version]
- Wen, B.; Zhou, R.; Feng, Q.; Wang, Q.; Wang, J.; Liu, S. IQuant: An automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 2014, 14, 2280–2285. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Bi, Z.; Dai, K.; Li, P.; Huang, R.; Wu, S.; Bao, W. A Functional Variant in the Aquaporin-3 Promoter Modulates Its Expression and Correlates with Resistance to Porcine Epidemic Virus Infection in Porcine Intestinal Epithelial Cells. Front. Microbiol. 2022, 13, 877644. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Hui, P.; Uemoto, Y.; Ding, Y.; Yin, Z.; Bao, W. Metabolomic and Proteomic Profiling of Porcine Intestinal Epithelial Cells Infected with Porcine Epidemic Diarrhea Virus. Int. J. Mol. Sci. 2023, 24, 5071. https://doi.org/10.3390/ijms24065071
Wang H, Hui P, Uemoto Y, Ding Y, Yin Z, Bao W. Metabolomic and Proteomic Profiling of Porcine Intestinal Epithelial Cells Infected with Porcine Epidemic Diarrhea Virus. International Journal of Molecular Sciences. 2023; 24(6):5071. https://doi.org/10.3390/ijms24065071
Chicago/Turabian StyleWang, Haifei, Peng Hui, Yoshinobu Uemoto, Yueyun Ding, Zongjun Yin, and Wenbin Bao. 2023. "Metabolomic and Proteomic Profiling of Porcine Intestinal Epithelial Cells Infected with Porcine Epidemic Diarrhea Virus" International Journal of Molecular Sciences 24, no. 6: 5071. https://doi.org/10.3390/ijms24065071
APA StyleWang, H., Hui, P., Uemoto, Y., Ding, Y., Yin, Z., & Bao, W. (2023). Metabolomic and Proteomic Profiling of Porcine Intestinal Epithelial Cells Infected with Porcine Epidemic Diarrhea Virus. International Journal of Molecular Sciences, 24(6), 5071. https://doi.org/10.3390/ijms24065071