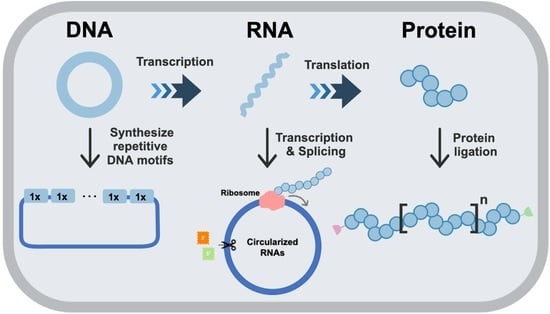
Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers
Abstract
:1. Introduction
2. Construction of Repetitive Genes for Repetitive Protein
2.1. Golden Gate DNA Assembly
2.2. Rolling Circle Amplification
2.3. Combinatorial Codon Scrambling for DNA Synthesis and Amplification
3. Expression of Repetitive Proteins from Circular mRNA
3.1. Advantageous in PBM Production Using cmRNAs
3.2. Examples of PBM Production from cmRNAs
3.3. Challenges and Opportunities in Producing PBMs from cmRNAs
4. Synthesis of Repetitive Proteins Using Protein Ligation or Polymerization
4.1. Ligation of the Protein Backbones via Peptide Bonds
4.1.1. Split Intein (SI)-Mediated Protein Ligation
4.1.2. Split Intein (SI)-Mediated Protein Polymerization
4.1.3. Sortase-Mediated Protein Ligation
4.2. Crosslinking of Proteins Using Side-Chain Chemistry
4.2.1. Catcher/Tag Reactions
4.2.2. Other Enzyme-Mediated Protein Conjugation Reactions
4.2.3. Cysteine-Based Protein Crosslinking
4.2.4. Lysine Side Chain Modification
4.2.5. Tyrosine Side Chain Oxidation
4.3. Comparing Different Protein Ligation and Conjugation Approaches to Synthesize Repetitive Protein Oligomers and Polymers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Freeman, R.; Boekhoven, J.; Dickerson, M.B.; Naik, R.R.; Stupp, S.I. Biopolymers and Supramolecular Polymers as Biomaterials for Biomedical Applications. MRS Bull. 2015, 40, 1089–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.; Dai, B.; Qiao, J.B.; Li, W.; Fortner, J.D.; Zhang, F. Microbially Synthesized Repeats of Mussel Foot Protein Display Enhanced Underwater Adhesion. ACS Appl. Mater. Interfaces 2018, 10, 43003–43012. [Google Scholar] [CrossRef] [PubMed]
- Bowen, C.H.; Dai, B.; Sargent, C.J.; Bai, W.; Ladiwala, P.; Feng, H.; Huang, W.; Kaplan, D.L.; Galazka, J.M.; Zhang, F. Recombinant Spidroins Fully Replicate Primary Mechanical Properties of Natural Spider Silk. Biomacromolecules 2018, 19, 3853–3860. [Google Scholar] [CrossRef] [Green Version]
- Lipońska, A.; Ousalem, F.; Aalberts, D.P.; Hunt, J.F.; Boël, G. The New Strategies to Overcome Challenges in Protein Production in Bacteria. Microb. Biotechnol. 2019, 12, 44–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer-Miralles, N.; Villaverde, A. Bacterial Cell Factories for Recombinant Protein Production; Expanding the Catalogue. Microb. Cell Factories 2013, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Omenetto, F.G.; Kaplan, D.L. New Opportunities for an Ancient Material. Science 2010, 329, 528–531. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Jeon, J.; Zhu, Y.; Hoppe, E.D.; Jun, Y.-S.; Genin, G.M.; Zhang, F. A Biosynthetic Hybrid Spidroin-Amyloid-Mussel Foot Protein for Underwater Adhesion on Diverse Surfaces. ACS Appl. Mater. Interfaces 2021, 13, 48457–48468. [Google Scholar] [CrossRef]
- Bowen, C.H.; Sargent, C.J.; Wang, A.; Zhu, Y.; Chang, X.; Li, J.; Mu, X.; Galazka, J.M.; Jun, Y.-S.; Keten, S.; et al. Microbial Production of Megadalton Titin Yields Fibers with Advantageous Mechanical Properties. Nat. Commun. 2021, 12, 5182. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Y.; Yu, H.; Dai, B.; Jun, Y.-S.; Zhang, F. Microbially Synthesized Polymeric Amyloid Fiber Promotes β-Nanocrystal Formation and Displays Gigapascal Tensile Strength. ACS Nano 2021, 15, 11843–11853. [Google Scholar] [CrossRef]
- Li, J.; Zhang, F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. Int. J. Mol. Sci. 2021, 22, 10698. [Google Scholar] [CrossRef]
- Desai, M.S.; Lee, S.-W. Protein-Based Functional Nanomaterial Design for Bioengineering Applications: Protein-Based Functional Nanomaterial Design. WIREs Nanomed. Nanobiotechnol. 2015, 7, 69–97. [Google Scholar] [CrossRef] [PubMed]
- Dinjaski, N.; Huang, W.; Kaplan, D.L. Recursive Directional Ligation Approach for Cloning Recombinant Spider Silks. Pept. Self-Assem. 2018, 1777, 181–192. [Google Scholar]
- Jung, H.; Pena-Francesch, A.; Saadat, A.; Sebastian, A.; Kim, D.H.; Hamilton, R.F.; Albert, I.; Allen, B.D.; Demirel, M.C. Molecular Tandem Repeat Strategy for Elucidating Mechanical Properties of High-Strength Proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 6478–6483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.-S.; Ryum, J.; Park, S.-Y.; Kim, B.-G.; Kim, D.-M.; Won, J.-I. A New Cloning Strategy for Generating Multiple Repeats of a Repetitive Polypeptide Based on Non-Template PCR. Biotechnol. Lett. 2011, 33, 977–983. [Google Scholar] [CrossRef]
- Moradali, M.F.; Rehm, B.H.A. Bacterial Biopolymers: From Pathogenesis to Advanced Materials. Nat. Rev. Microbiol. 2020, 18, 195–210. [Google Scholar] [CrossRef]
- Xu, J.; Dong, Q.; Yu, Y.; Niu, B.; Ji, D.; Li, M.; Huang, Y.; Chen, X.; Tan, A. Mass Spider Silk Production through Targeted Gene Replacement in Bombyx Mori. Proc. Natl. Acad. Sci. USA 2018, 115, 8757–8762. [Google Scholar] [CrossRef] [Green Version]
- Dai, B.; Sargent, C.J.; Gui, X.; Liu, C.; Zhang, F. Fibril Self-Assembly of Amyloid–Spider Silk Block Polypeptides. Biomacromolecules 2019, 20, 2015–2023. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.C.; Chilkoti, A. Combinatorial Codon Scrambling Enables Scalable Gene Synthesis and Amplification of Repetitive Proteins. Nat. Mater. 2016, 15, 419–424. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, P.; Zhao, D.; Zhu, L.; Tang, J.; Leng, W.; Su, J.; Liu, Y.; Bi, C.; Zhang, X. Engineering Circularized MRNAs for the Production of Spider Silk Proteins. Appl. Environ. Microbiol. 2022, 88, e00028-22. [Google Scholar] [CrossRef]
- Lee, S.O.; Xie, Q.; Fried, S.D. Optimized Loopable Translation as a Platform for the Synthesis of Repetitive Proteins. ACS Cent. Sci. 2021, 7, 1736–1750. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An Emerging Frontier in RNA Therapeutic Targets, RNA Therapeutics, and MRNA Vaccines. J. Control. Release 2022, 348, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, Y.; Wang, X.; Shen, J.; An, W. Advances in Circular RNA and Its Applications. Int. J. Med. Sci. 2022, 19, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.H.; Muir, T.W. Inteins: Nature’s Gift to Protein Chemists. Chem. Sci. 2013, 5, 446–461. [Google Scholar] [CrossRef] [Green Version]
- Züger, S.; Iwai, H. Intein-Based Biosynthetic Incorporation of Unlabeled Protein Tags into Isotopically Labeled Proteins for NMR Studies. Nat. Biotechnol. 2005, 23, 736–740. [Google Scholar] [CrossRef]
- Bai, W.; Sargent, C.J.; Choi, J.-M.; Pappu, R.V.; Zhang, F. Covalently-Assembled Single-Chain Protein Nanostructures with Ultra-High Stability. Nat. Commun. 2019, 10, 3317. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.-X.; Qian, Z.-G.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-Sized Recombinant Spider Silk Protein Produced in Metabolically Engineered Escherichia Coli Results in a Strong Fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Qin, X.; Qiao, J.B.; Zeng, Q.; Fortner, J.D.; Zhang, F. Graphene Oxide/Mussel Foot Protein Composites for High-Strength and Ultra-Tough Thin Films. Sci. Rep. 2020, 10, 19082. [Google Scholar] [CrossRef]
- Bowen, C.H.; Reed, T.J.; Sargent, C.J.; Mpamo, B.; Galazka, J.M.; Zhang, F. Seeded Chain-Growth Polymerization of Proteins in Living Bacterial Cells. ACS Synth. Biol. 2019, 8, 2651–2658. [Google Scholar] [CrossRef]
- Antos, J.M.; Truttmann, M.C.; Ploegh, H.L. Recent Advances in Sortase-Catalyzed Ligation Methodology. Curr. Opin. Struct. Biol. 2016, 38, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Domeradzka, N.E.; Werten, M.W.; de Wolf, F.A.; de Vries, R. Protein Cross-Linking Tools for the Construction of Nanomaterials. Curr. Opin. Biotechnol. 2016, 39, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Pishesha, N.; Ingram, J.R.; Ploegh, H.L. Sortase A: A Model for Transpeptidation and Its Biological Applications. Annu. Rev. Cell Dev. Biol. 2018, 34, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Ishikawa, S.; Nagamune, T. Design of Ca2+-Independent Staphylococcus Aureus Sortase A Mutants. Biotechnol. Bioeng. 2012, 109, 2955–2961. [Google Scholar] [CrossRef]
- Witte, M.D.; Wu, T.; Guimaraes, C.P.; Theile, C.S.; Blom, A.E.M.; Ingram, J.R.; Li, Z.; Kundrat, L.; Goldberg, S.D.; Ploegh, H.L. Site-Specific Protein Modification Using Immobilized Sortase in Batch and Continuous-Flow Systems. Nat. Protoc. 2015, 10, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Warden-Rothman, R.; Caturegli, I.; Popik, V.; Tsourkas, A. Sortase-Tag Expressed Protein Ligation: Combining Protein Purification and Site-Specific Bioconjugation into a Single Step. Anal. Chem. 2013, 85, 11090–11097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamura, Y.; Hirakawa, H.; Yamaguchi, S.; Nagamune, T. Enhancement of Sortase A-Mediated Protein Ligation by Inducing a β-Hairpin Structure around the Ligation Site. Chem. Commun. 2011, 47, 4742–4744. [Google Scholar] [CrossRef]
- Zhang, J.; Yamaguchi, S.; Hirakawa, H.; Nagamune, T. Intracellular Protein Cyclization Catalyzed by Exogenously Transduced Streptococcus Pyogenes Sortase A. J. Biosci. Bioeng. 2013, 116, 298–301. [Google Scholar] [CrossRef]
- Samantaray, S.; Marathe, U.; Dasgupta, S.; Nandicoori, V.K.; Roy, R.P. Peptide−Sugar Ligation Catalyzed by Transpeptidase Sortase: A Facile Approach to Neoglycoconjugate Synthesis. J. Am. Chem. Soc. 2008, 130, 2132–2133. [Google Scholar] [CrossRef]
- Bierlmeier, J.; Álvaro-Benito, M.; Scheffler, M.; Sturm, K.; Rehkopf, L.; Freund, C.; Schwarzer, D. Sortase-Mediated Multi-Fragment Assemblies by Ligation Site Switching. Angew. Chem. Int. Ed. 2022, 61, e202109032. [Google Scholar] [CrossRef]
- Keeble, A.H.; Yadav, V.K.; Ferla, M.P.; Bauer, C.C.; Chuntharpursat-Bon, E.; Huang, J.; Bon, R.S.; Howarth, M. DogCatcher Allows Loop-Friendly Protein-Protein Ligation. Cell Chem. Biol. 2022, 29, 339–350.e10. [Google Scholar] [CrossRef]
- Zhang, W.-B.; Sun, F.; Tirrell, D.A.; Arnold, F.H. Controlling Macromolecular Topology with Genetically Encoded SpyTag–SpyCatcher Chemistry. J. Am. Chem. Soc. 2013, 135, 13988–13997. [Google Scholar] [CrossRef] [Green Version]
- Keeble, A.H.; Howarth, M. Power to the Protein: Enhancing and Combining Activities Using the Spy Toolbox. Chem. Sci. 2020, 11, 7281–7291. [Google Scholar] [CrossRef] [PubMed]
- Khairil Anuar, I.N.A.; Banerjee, A.; Keeble, A.H.; Carella, A.; Nikov, G.I.; Howarth, M. Spy&Go Purification of SpyTag-Proteins Using Pseudo-SpyCatcher to Access an Oligomerization Toolbox. Nat. Commun. 2019, 10, 1734. [Google Scholar] [PubMed] [Green Version]
- Yi, Q.; Dai, X.; Park, B.M.; Gu, J.; Luo, J.; Wang, R.; Yu, C.; Kou, S.; Huang, J.; Lakerveld, R.; et al. Directed Assembly of Genetically Engineered Eukaryotic Cells into Living Functional Materials via Ultrahigh-Affinity Protein Interactions. Sci. Adv. 2022, 8, eade0073. [Google Scholar] [CrossRef] [PubMed]
- Fok, H.K.F.; Yang, Z.; Jiang, B.; Sun, F. From 4-Arm Star Proteins to Diverse Stimuli-Responsive Molecular Networks Enabled by Orthogonal Genetically Encoded Click Chemistries. Polym. Chem. 2022, 13, 2331–2339. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Yang, Z.; Luo, J.; Tian, X.; Liu, K.; Kou, S.; Sun, F. Versatile Engineered Protein Hydrogels Enabling Decoupled Mechanical and Biochemical Tuning for Cell Adhesion and Neurite Growth. ACS Appl. Nano Mater. 2018, 1, 1579–1585. [Google Scholar] [CrossRef]
- Reddington, S.C.; Howarth, M. Secrets of a Covalent Interaction for Biomaterials and Biotechnology: SpyTag and SpyCatcher. Curr. Opin. Chem. Biol. 2015, 29, 94–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, J.O.; Veggiani, G.; Howarth, M. SpyLigase Peptide–Peptide Ligation Polymerizes Affibodies to Enhance Magnetic Cancer Cell Capture. Proc. Natl. Acad. Sci. USA 2014, 111, E1176–E1181. [Google Scholar] [CrossRef] [Green Version]
- Krüger, T.; Dierks, T.; Sewald, N. Formylglycine-Generating Enzymes for Site-Specific Bioconjugation. Biol. Chem. 2019, 400, 289–297. [Google Scholar] [CrossRef]
- Hudak, J.E.; Barfield, R.M.; de Hart, G.W.; Grob, P.; Nogales, E.; Bertozzi, C.R.; Rabuka, D. Synthesis of Heterobifunctional Protein Fusions Using Copper-Free Click Chemistry and the Aldehyde Tag. Angew. Chem. Int. Ed. 2012, 51, 4161–4165. [Google Scholar] [CrossRef]
- Baalmann, M.; Best, M.; Wombacher, R. Site-Specific Protein Labeling Utilizing Lipoic Acid Ligase (LplA) and Bioorthogonal Inverse Electron Demand Diels-Alder Reaction. In Methods in Molecular Biology; Lemke, E.A., Ed.; Springer: New York, NY, USA, 2018; Volume 1728, pp. 365–387. [Google Scholar]
- Hofmann, R.; Akimoto, G.; Wucherpfennig, T.G.; Zeymer, C.; Bode, J.W. Lysine Acylation Using Conjugating Enzymes for Site-Specific Modification and Ubiquitination of Recombinant Proteins. Nat. Chem. 2020, 12, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Ravasco, J.M.J.M.; Faustino, H.; Trindade, A.; Gois, P.M.P. Bioconjugation with Maleimides: A Useful Tool for Chemical Biology. Chem. Eur. J. 2019, 25, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Du, W.; Xiong, C.-Y.; DeNardo, G.L.; DeNardo, S.J.; Gervay-Hague, J. Construction of Di-ScFv through a Trivalent Alkyne—Azide 1,3-Dipolar Cycloaddition. Chem. Commun. 2007, 43, 695–697. [Google Scholar] [CrossRef]
- Taylor, R.J.; Geeson, M.B.; Journeaux, T.; Bernardes, G.J.L. Chemical and Enzymatic Methods for Post-Translational Protein–Protein Conjugation. J. Am. Chem. Soc. 2022, 144, 14404–14419. [Google Scholar] [CrossRef]
- Azevedo, C.; Saiardi, A. Why Always Lysine? The Ongoing Tale of One of the Most Modified Amino Acids. Adv. Biol. Regul. 2016, 60, 144–150. [Google Scholar] [CrossRef]
- Slusarewicz, P.; Zhu, K.; Hedman, T. Kinetic Characterization and Comparison of Various Protein Crosslinking Reagents for Matrix Modification. J. Mater. Sci. Mater. Med. 2010, 21, 1175–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Wu, H.; Schnier, P.D.; Liu, Y.; Liu, J.; Wang, N.; DeGrado, W.F.; Wang, L. Proximity-Enhanced SuFEx Chemical Cross-Linker for Specific and Multitargeting Cross-Linking Mass Spectrometry. Proc. Natl. Acad. Sci. USA 2018, 115, 11162–11167. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Cai, L.; Sun, W.; Cheng, R.; Wang, N.; Jin, L.; Rozovsky, S.; Seiple, I.B.; Wang, L. Photocaged Quinone Methide Crosslinkers for Light-Controlled Chemical Crosslinking of Protein–Protein and Protein–DNA Complexes. Angew. Chem. Int. Ed. 2019, 58, 18839–18843. [Google Scholar] [CrossRef]
- Lobba, M.J.; Fellmann, C.; Marmelstein, A.M.; Maza, J.C.; Kissman, E.N.; Robinson, S.A.; Staahl, B.T.; Urnes, C.; Lew, R.J.; Mogilevsky, C.S.; et al. Site-Specific Bioconjugation through Enzyme-Catalyzed Tyrosine–Cysteine Bond Formation. ACS Cent. Sci. 2020, 6, 1564–1571. [Google Scholar] [CrossRef]
- Permana, D.; Minamihata, K.; Goto, M.; Kamiya, N. Laccase-Catalyzed Bioconjugation of Tyrosine-Tagged Functional Proteins. J. Biosci. Bioeng. 2018, 126, 559–566. [Google Scholar] [CrossRef]
- Stengl, A.; Gerlach, M.; Kasper, M.-A.; Hackenberger, C.P.R.; Leonhardt, H.; Schumacher, D.; Helma, J. TuPPL: Tub-Tag Mediated C-Terminal Protein–Protein-Ligation Using Complementary Click-Chemistry Handles. Org. Biomol. Chem. 2019, 17, 4964–4969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, R.; Hakanpää, J.; Elfving, K.; Taberman, H.; Linder, M.B.; Aranko, A.S. Biomolecular Click Reactions Using a Minimal PH-Activated Catcher/Tag Pair for Producing Native-Sized Spider-Silk Proteins. Angew. Chem. Int. Ed. 2023, 62, e202216371. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Gu, P.; Zhang, F. Steps towards ‘Drop-in’ Biofuels: Focusing on Metabolic Pathways. Curr. Opin. Biotechnol. 2018, 53, 26–32. [Google Scholar] [CrossRef]
- Bai, W.; Geng, W.; Wang, S.; Zhang, F. Biosynthesis, Regulation, and Engineering of Microbially Produced Branched Biofuels. Biotechnol. Biofuels 2019, 12, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Qiao, J.B.; Bentley, G.J.; Liu, D.; Zhang, F. Modular Pathway Engineering for the Microbial Production of Branched-Chain Fatty Alcohols. Biotechnol. Biofuels 2017, 10, 244. [Google Scholar] [CrossRef]
- Bai, W.; Anthony, W.E.; Hartline, C.J.; Wang, S.; Wang, B.; Ning, J.; Hsu, F.-F.; Dantas, G.; Zhang, F. Engineering Diverse Fatty Acid Compositions of Phospholipids in Escherichia Coli. Metab. Eng. 2022, 74, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.C.; Hartline, C.J.; Zhang, F. Engineering Microbial Metabolite Dynamics and Heterogeneity. Biotechnol. J. 2017, 12, 1700422. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, F. Control Strategies to Manage Trade-Offs during Microbial Production. Curr. Opin. Biotechnol. 2020, 66, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, F. Metabolic Feedback Circuits Provide Rapid Control of Metabolite Dynamics. ACS Synth. Biol. 2018, 7, 347–356. [Google Scholar] [CrossRef]





| Golden Gate Assembly | Rolling Circle Amplification | Codon Scrambling | Circular mRNA | |
|---|---|---|---|---|
| Unwanted DNA between monomers | No | Yes | No | Yes |
| Precise control of repeat numbers | Yes | No | Yes | No |
| Time consuming | + | ++ | + | + |
| Split Intein (SI) -Mediated Protein Ligation | Split Intein (SI)-Mediated Protein Polymerization | Sortase-Mediated Protein Ligation | |
|---|---|---|---|
| Ligation rate | ++ | ++ | + |
| Ligation efficiency | ++ | + | |
| High MW protein yield | + | ++ |
| Catcher/Tag Reactions | Cysteine-Based Protein Crosslinking | Lysine Side Chain Modification | Tyrosine Side Chain Oxidation | |
|---|---|---|---|---|
| Reaction rate | ++ | + | + | + |
| Conjugation yield | + | ++ | ++ | + |
| Have large conjugation domain left | Yes | No | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, J.; Subramani, S.V.; Lee, K.Z.; Jiang, B.; Zhang, F. Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers. Int. J. Mol. Sci. 2023, 24, 6416. https://doi.org/10.3390/ijms24076416
Jeon J, Subramani SV, Lee KZ, Jiang B, Zhang F. Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers. International Journal of Molecular Sciences. 2023; 24(7):6416. https://doi.org/10.3390/ijms24076416
Chicago/Turabian StyleJeon, Juya, Shri Venkatesh Subramani, Kok Zhi Lee, Bojing Jiang, and Fuzhong Zhang. 2023. "Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers" International Journal of Molecular Sciences 24, no. 7: 6416. https://doi.org/10.3390/ijms24076416
APA StyleJeon, J., Subramani, S. V., Lee, K. Z., Jiang, B., & Zhang, F. (2023). Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers. International Journal of Molecular Sciences, 24(7), 6416. https://doi.org/10.3390/ijms24076416