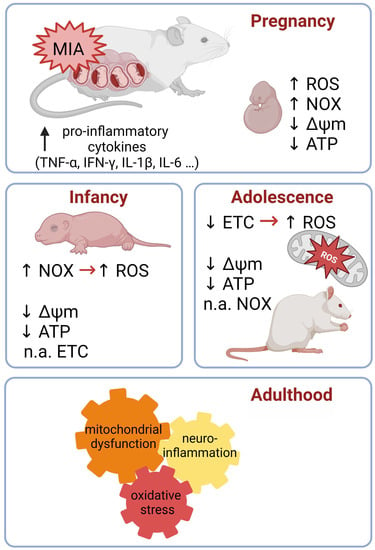
Developmental Stage-Dependent Changes in Mitochondrial Function in the Brain of Offspring Following Prenatal Maternal Immune Activation
Abstract
:1. Introduction
2. Results
2.1. Maternal Immune Activation Induces Free Radical Generation and Mitochondrial Dysfunction in Fetuses 24 h after the Administration of LPS to a Pregnant Mother
2.2. The Impact of MIA on the Oxidative Stress and Function of Mitochondria in the Brain of 7-Day-Old Offspring
2.3. MIA Alters Gene Expression for the Fusion and Fission Proteins in the Brain of 7-Day-Old Offspring
2.4. No Effect of MIA on the Mitochondria Content in the Brain of 7-Day-Old Offspring
2.5. The Impact of MIA on the Oxidative Stress and Mitochondrial Function in the Cerebral Cortex of Adolescent 54-Day-Old Offspring
2.6. Mitochondrial Dynamics: Fission and Fusion Processes in Response to MIA in the Cerebral Cortex of Adolescent 54-Day-Old Offspring
2.7. The Influence of MIA on the Mitochondria Content in the Cerebral Cortex of Adolescent 54-Day-Old Offspring
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Animals and Experimental Procedure
4.3. Mitochondria Isolation and Determination of the Mitochondrial Membrane Potential (ΔΨm)
4.4. Measurement of the Reactive Oxygen Species (ROS) Level
4.5. Measurement of the Superoxide Radical Level
4.6. Measurement of the NOX/NADPH Oxidase (NOX) Activity
4.7. Determination of Mitochondrial ATP Levels
4.8. Determination of Gene Expression (Real-Time PCR)
4.9. Citrate Synthase Activity Assay
4.10. Determination of Protein Level
4.11. Immunochemical Determination of Protein Levels (Western Blot Analysis)
4.12. Electron Transport Chain Complexes Activity Assays
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013; pp. 591–643. [Google Scholar] [CrossRef]
- Chiarotti, F.; Venerosi, A. Epidemiology of Autism Spectrum Disorders: A Review of Worldwide Prevalence Estimates Since 2014. Brain Sci. 2020, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Elsabbagh, M.; Divan, G.; Koh, Y.J.; Kim, Y.S.; Kauchali, S.; Marcín, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. Off. J. Int. Soc. Autism Res. 2012, 5, 160–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyall, K.; Croen, L.; Daniels, J.; Fallin, M.D.; Ladd-Acosta, C.; Lee, B.K.; Park, B.Y.; Snyder, N.W.; Schendel, D.; Volk, H.; et al. The Changing Epidemiology of Autism Spectrum Disorders. Annu. Rev. Public Health 2017, 38, 81–102. [Google Scholar] [CrossRef] [Green Version]
- Zuckerman, L.; Rehavi, M.; Nachman, R.; Weiner, I. Immune activation during pregnancy in rats leads to a postpubertal emergence of disrupted latent inhibition, dopaminergic hyperfunction, and altered limbic morphology in the offspring: A novel neurodevelopmental model of schizophrenia. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2003, 28, 1778–1789. [Google Scholar] [CrossRef] [Green Version]
- Careaga, M.; Murai, T.; Bauman, M.D. Maternal Immune Activation and Autism Spectrum Disorder: From Rodents to Nonhuman and Human Primates. Biol. Psychiatry 2017, 81, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Parker-Athill, E.C.; Tan, J. Maternal immune activation and autism spectrum disorder: Interleukin-6 signaling as a key mechanistic pathway. Neuro-Signals 2010, 18, 113–128. [Google Scholar] [CrossRef] [Green Version]
- Lombardo, M.V.; Moon, H.M.; Su, J.; Palmer, T.D.; Courchesne, E.; Pramparo, T. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol. Psychiatry 2018, 23, 1001–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knuesel, I.; Chicha, L.; Britschgi, M.; Schobel, S.A.; Bodmer, M.; Hellings, J.A.; Toovey, S.; Prinssen, E.P. Maternal immune activation and abnormal brain development across CNS disorders. Nat. Rev. Neurol. 2014, 10, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.B.; Yim, Y.S.; Wong, H.; Kim, S.; Kim, H.; Kim, S.V.; Hoeffer, C.A.; Littman, D.R.; Huh, J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016, 351, 933–939. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.; Hoeffer, C. Maternal IL-17A in autism. Exp. Neurol. 2018, 299, 228–240. [Google Scholar] [CrossRef]
- Yasumatsu, K.; Nagao, J.I.; Arita-Morioka, K.I.; Narita, Y.; Tasaki, S.; Toyoda, K.; Ito, S.; Kido, H.; Tanaka, Y. Bacterial-induced maternal interleukin-17A pathway promotes autistic-like behaviors in mouse offspring. Exp. Anim. 2020, 69, 250–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.E.; Li, J.; Garbett, K.; Mirnics, K.; Patterson, P.H. Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 10695–10702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, E.Y.; Patterson, P.H. Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav. Immun. 2011, 25, 604–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Zheng, Y.; Liu, Y.; Zhang, X.; Zhao, J. Minocycline alleviates behavioral deficits and inhibits microglial activation in the offspring of pregnant mice after administration of polyriboinosinic–polyribocytidilic acid. Psychiatry Res. 2014, 219, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Van den Eynde, K.; Missault, S.; Fransen, E.; Raeymaekers, L.; Willems, R.; Drinkenburg, W.; Timmermans, J.-P.; Kumar-Singh, S.; Dedeurwaerdere, S. Hypolocomotive behaviour associated with increased microglia in a prenatal immune activation model with relevance to schizophrenia. Behav. Brain Res. 2014, 258, 179–186. [Google Scholar] [CrossRef]
- Cieślik, M.; Gąssowska-Dobrowolska, M.; Jęśko, H.; Czapski, G.A.; Wilkaniec, A.; Zawadzka, A.; Dominiak, A.; Polowy, R.; Filipkowski, R.K.; Boguszewski, P.M.; et al. Maternal Immune Activation Induces Neuroinflammation and Cortical Synaptic Deficits in the Adolescent Rat Offspring. Int. J. Mol. Sci. 2020, 21, 4097. [Google Scholar] [CrossRef]
- Rossignol, D.A.; Frye, R.E. A review of research trends in physiological abnormalities in autism spectrum disorders: Immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol. Psychiatry 2012, 17, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Frye, R.E.; Cakir, J.; Rose, S.; Palmer, R.F.; Austin, C.; Curtin, P.; Arora, M. Mitochondria May Mediate Prenatal Environmental Influences in Autism Spectrum Disorder. J. Pers. Med. 2021, 11, 218. [Google Scholar] [CrossRef]
- Tang, G.; Gutierrez Rios, P.; Kuo, S.-H.; Akman, H.O.; Rosoklija, G.; Tanji, K.; Dwork, A.; Schon, E.A.; DiMauro, S.; Goldman, J.; et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol. Dis. 2013, 54, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, A.V.; Margreiter, R. Heterogeneity of Mitochondria and Mitochondrial Function within Cells as Another Level of Mitochondrial Complexity. Int. J. Mol. Sci. 2009, 10, 1911–1929. [Google Scholar] [CrossRef] [Green Version]
- Reczek, C.R.; Chandel, N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015, 33, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, M.D.; Orr, A.L.; Perevoshchikova, I.V.; Quinlan, C.L. The role of mitochondrial function and cellular bioenergetics in ageing and disease. Br. J. Dermatol. 2013, 169 (Suppl. 2), 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, D.C.; Fan, W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion 2010, 10, 12–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxidative Med. Cell. Longev. 2019, 2019, 6175804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Fang, P.; Mai, J.; Choi, E.T.; Wang, H.; Yang, X.-f. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013, 6, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobley, J.N.; Fiorello, M.L.; Bailey, D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018, 15, 490–503. [Google Scholar] [CrossRef]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Guo, C.; Sun, L.; Chen, X.; Zhang, D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 2013, 8, 2003–2014. [Google Scholar] [CrossRef]
- Hollensworth, S.B.; Shen, C.; Sim, J.E.; Spitz, D.R.; Wilson, G.L.; LeDoux, S.P. Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic. Biol. Med. 2000, 28, 1161–1174. [Google Scholar] [CrossRef]
- Rossignol, D.A.; Frye, R.E. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol. Psychiatry 2012, 17, 290–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pons, R.; Andreu, A.L.; Checcarelli, N.; Vilà, M.R.; Engelstad, K.; Sue, C.M.; Shungu, D.; Haggerty, R.; de Vivo, D.C.; DiMauro, S. Mitochondrial DNA abnormalities and autistic spectrum disorders. J. Pediatr. 2004, 144, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Gu, F.; Essa, M.M.; Wegiel, J.; Kaur, K.; Brown, W.T.; Chauhan, V. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J. Neurochem. 2011, 117, 209–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, A.; Audhya, T.; Chauhan, V. Brain region-specific glutathione redox imbalance in autism. Neurochem. Res. 2012, 37, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Anitha, A.; Nakamura, K.; Thanseem, I.; Yamada, K.; Iwayama, Y.; Toyota, T.; Matsuzaki, H.; Miyachi, T.; Yamada, S.; Tsujii, M.; et al. Brain region-specific altered expression and association of mitochondria-related genes in autism. Mol. Autism 2012, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Rose, S.; Melnyk, S.; Pavliv, O.; Bai, S.; Nick, T.G.; Frye, R.E.; James, S.J. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Transl. Psychiatry 2012, 2, e134. [Google Scholar] [CrossRef] [Green Version]
- Rose, S.; Niyazov, D.M.; Rossignol, D.A.; Goldenthal, M.; Kahler, S.G.; Frye, R.E. Clinical and Molecular Characteristics of Mitochondrial Dysfunction in Autism Spectrum Disorder. Mol. Diagn. Ther. 2018, 22, 571–593. [Google Scholar] [CrossRef] [Green Version]
- Frye, R.E. Mitochondrial Dysfunction in Autism Spectrum Disorder: Unique Abnormalities and Targeted Treatments. Semin. Pediatr. Neurol. 2020, 35, 100829. [Google Scholar] [CrossRef]
- Frye, R.E.; Rossignol, D.A. Mitochondrial Dysfunction Can Connect the Diverse Medical Symptoms Associated With Autism Spectrum Disorders. Pediatr. Res. 2011, 69, 41–47. [Google Scholar] [CrossRef]
- Wang, Y.; Picard, M.; Gu, Z. Genetic Evidence for Elevated Pathogenicity of Mitochondrial DNA Heteroplasmy in Autism Spectrum Disorder. PLoS Genet. 2016, 12, e1006391. [Google Scholar] [CrossRef] [Green Version]
- Varga, N.Á.; Pentelényi, K.; Balicza, P.; Gézsi, A.; Reményi, V.; Hársfalvi, V.; Bencsik, R.; Illés, A.; Prekop, C.; Molnár, M.J. Mitochondrial dysfunction and autism: Comprehensive genetic analyses of children with autism and mtDNA deletion. Behav. Brain Funct. 2018, 14, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, G.; Diogo, L.; Grazina, M.; Garcia, P.; Ataíde, A.; Marques, C.; Miguel, T.; Borges, L.; Vicente, A.M.; Oliveira, C.R. Mitochondrial dysfunction in autism spectrum disorders: A population-based study. Dev. Med. Child Neurol. 2005, 47, 185–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Xu, M.; Xo, R.; Mates, A.; Wilson, G.; Pearsall IV, A.; Grishko, V. Mitochondrial DNA damage is involved in apoptosis caused by pro-inflammatory cytokines in human OA chondrocytes. Osteoarthr. Cartil. 2010, 18, 424–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.; Berk, M. The many roads to mitochondrial dysfunction in neuroimmune and neuropsychiatric disorders. BMC Med. 2015, 13, 68. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.; Maes, M. Mitochondrial dysfunctions in myalgic encephalomyelitis/chronic fatigue syndrome explained by activated immuno-inflammatory, oxidative and nitrosative stress pathways. Metab. Brain Dis. 2014, 29, 19–36. [Google Scholar] [CrossRef]
- Newman, L.E.; Shadel, G.S. Pink1/Parkin link inflammation, mitochondrial stress, and neurodegeneration. J. Cell Biol. 2018, 217, 3327–3329. [Google Scholar] [CrossRef] [Green Version]
- Dominiak, A.; Wilkaniec, A.; Jęśko, H.; Czapski, G.A.; Lenkiewicz, A.M.; Kurek, E.; Wroczyński, P.; Adamczyk, A. Selol, an organic selenium donor, prevents lipopolysaccharide-induced oxidative stress and inflammatory reaction in the rat brain. Neurochem. Int. 2017, 108, 66–77. [Google Scholar] [CrossRef]
- Cieślik, M.; Gassowska-Dobrowolska, M.; Zawadzka, A.; Frontczak-Baniewicz, M.; Gewartowska, M.; Dominiak, A.; Czapski, G.A.; Adamczyk, A. The Synaptic Dysregulation in Adolescent Rats Exposed to Maternal Immune Activation. Front. Mol. Neurosci. 2021, 13, 555290. [Google Scholar] [CrossRef]
- Nadeem, A.; Ahmad, S.F.; Bakheet, S.A.; Al-Harbi, N.O.; Al-Ayadhi, L.Y.; Attia, S.M.; Zoheir, K.M.A. Toll-like receptor 4 signaling is associated with upregulated NADPH oxidase expression in peripheral T cells of children with autism. Brain Behav. Immun. 2017, 61, 146–154. [Google Scholar] [CrossRef]
- Eigentler, A.; Draxl, A.; Wiethüchter, A.; Kuznetsov, A.; Lassing, B.; Gnaiger, E. Laboratory protocol: Citrate synthase, a mitochondrial marker enzyme. Mitochondrial Physiol. Netw. 2015, 17, 1–11. [Google Scholar]
- Larsen, S.; Nielsen, J.; Hansen, C.N.; Nielsen, L.B.; Wibrand, F.; Stride, N.; Schroder, H.D.; Boushel, R.; Helge, J.W.; Dela, F. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012, 590, 3349–3360. [Google Scholar] [CrossRef] [PubMed]
- Cenini, G.; Lloret, A.; Cascella, R. Oxidative Stress in Neurodegenerative Diseases: From a Mitochondrial Point of View. Oxidative Med. Cell. Longev. 2019, 2019, 2105607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pangrazzi, L.; Balasco, L.; Bozzi, Y. Oxidative Stress and Immune System Dysfunction in Autism Spectrum Disorders. Int. J. Mol. Sci. 2020, 21, 3293. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, X.-J.; Liu, H.; Mao, X.; Gui, L.-N.; Wang, H.; Cheng, Y. Oxidative stress marker aberrations in children with autism spectrum disorder: A systematic review and meta-analysis of 87 studies (N = 9109). Transl. Psychiatry 2021, 11, 15. [Google Scholar] [CrossRef]
- Abruzzo, P.M.; Matté, A.; Bolotta, A.; Federti, E.; Ghezzo, A.; Guarnieri, T.; Marini, M.; Posar, A.; Siciliano, A.; De Franceschi, L.; et al. Plasma peroxiredoxin changes and inflammatory cytokines support the involvement of neuro-inflammation and oxidative stress in Autism Spectrum Disorder. J. Transl. Med. 2019, 17, 332. [Google Scholar] [CrossRef] [Green Version]
- Lanté, F.; Meunier, J.; Guiramand, J.; Maurice, T.; Cavalier, M.; de Jesus Ferreira, M.-C.; Aimar, R.; Cohen-Solal, C.; Vignes, M.; Barbanel, G. Neurodevelopmental damage after prenatal infection: Role of oxidative stress in the fetal brain. Free Radic. Biol. Med. 2007, 42, 1231–1245. [Google Scholar] [CrossRef]
- Van Houten, B.; Woshner, V.; Santos, J.H. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair 2006, 5, 145–152. [Google Scholar] [CrossRef]
- Simões, L.R.; Sangiogo, G.; Tashiro, M.H.; Generoso, J.S.; Faller, C.J.; Dominguini, D.; Mastella, G.A.; Scaini, G.; Giridharan, V.V.; Michels, M.; et al. Maternal immune activation induced by lipopolysaccharide triggers immune response in pregnant mother and fetus, and induces behavioral impairment in adult rats. J. Psychiatr. Res. 2018, 100, 71–83. [Google Scholar] [CrossRef]
- Wang, X.; Yang, J.; Zhang, H.; Yu, J.; Yao, Z. Oral probiotic administration during pregnancy prevents autism-related behaviors in offspring induced by maternal immune activation via anti-inflammation in mice. Autism Res. 2019, 12, 576–588. [Google Scholar] [CrossRef]
- Vuillermot, S.; Luan, W.; Meyer, U.; Eyles, D. Vitamin D treatment during pregnancy prevents autism-related phenotypes in a mouse model of maternal immune activation. Mol. Autism 2017, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Oskvig, D.B.; Elkahloun, A.G.; Johnson, K.R.; Phillips, T.M.; Herkenham, M. Maternal immune activation by LPS selectively alters specific gene expression profiles of interneuron migration and oxidative stress in the fetus without triggering a fetal immune response. Brain Behav. Immun. 2012, 26, 623–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terzi, A.; Suter, D.M. The role of NADPH oxidases in neuronal development. Free Radic. Biol. Med. 2020, 154, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Brandes, R.P.; Weissmann, N.; Schröder, K. Nox family NADPH oxidases: Molecular mechanisms of activation. Free Radic. Biol. Med. 2014, 76, 208–226. [Google Scholar] [CrossRef] [PubMed]
- Nayernia, Z.; Jaquet, V.; Krause, K.H. New insights on NOX enzymes in the central nervous system. Antioxid. Redox Signal. 2014, 20, 2815–2837. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ibi, M.; Haga, R.; Iwata, K.; Matsumoto, M.; Asaoka, N.; Liu, J.; Katsuyama, M.; Yabe-Nishimura, C. NOX1/NADPH oxidase affects the development of autism-like behaviors in a maternal immune activation model. Biochem. Biophys. Res. Commun. 2021, 534, 59–66. [Google Scholar] [CrossRef]
- Nadeem, A.; Ahmad, S.F.; Attia, S.M.; Al-Ayadhi, L.Y.; Bakheet, S.A.; Al-Harbi, N.O. Oxidative and inflammatory mediators are upregulated in neutrophils of autistic children: Role of IL-17A receptor signaling. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 90, 204–211. [Google Scholar] [CrossRef]
- Schiavone, S.; Neri, M.; Trabace, L.; Turillazzi, E. The NADPH oxidase NOX2 mediates loss of parvalbumin interneurons in traumatic brain injury: Human autoptic immunohistochemical evidence. Sci. Rep. 2017, 7, 8752. [Google Scholar] [CrossRef] [Green Version]
- Gyllenhammer, L.E.; Entringer, S.; Buss, C.; Wadhwa, P.D. Developmental programming of mitochondrial biology: A conceptual framework and review. Proc. R. Soc. B Biol. Sci. 2020, 287, 20192713. [Google Scholar] [CrossRef]
- Hagberg, H.; Mallard, C.; Rousset, C.I.; Thornton, C. Mitochondria: Hub of injury responses in the developing brain. Lancet Neurol. 2014, 13, 217–232. [Google Scholar] [CrossRef]
- Lee, H.; Jose, P.A. Coordinated Contribution of NADPH Oxidase- and Mitochondria-Derived Reactive Oxygen Species in Metabolic Syndrome and Its Implication in Renal Dysfunction. Front. Pharmacol. 2021, 12, 670076. [Google Scholar] [CrossRef]
- Zhao, R.Z.; Jiang, S.; Zhang, L.; Yu, Z.B. Mitochondrial electron transport chain, ROS generation and uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadenas, S. Mitochondrial uncoupling, ROS generation and cardioprotection. Biochim. Biophys. Acta (BBA)—Bioenerg. 2018, 1859, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, B.S.; Freire-Brito, L.; Carrageta, D.F.; Oliveira, P.F.; Alves, M.G. Mitochondrial Uncoupling Proteins (UCPs) as Key Modulators of ROS Homeostasis: A Crosstalk between Diabesity and Male Infertility? Antioxidants 2021, 10, 1746. [Google Scholar] [CrossRef] [PubMed]
- Čater, M.; Križančić Bombek, L. Protective Role of Mitochondrial Uncoupling Proteins against Age-Related Oxidative Stress in Type 2 Diabetes Mellitus. Antioxidants 2022, 11, 1473. [Google Scholar] [CrossRef]
- Khacho, M.; Slack, R.S. Mitochondrial dynamics in the regulation of neurogenesis: From development to the adult brain. Dev. Dyn. 2018, 247, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleier, L.; Dröse, S. Superoxide generation by complex III: From mechanistic rationales to functional consequences. Biochim. Biophys. Acta 2013, 1827, 1320–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, J.; Fleming, A.M.; Xu, J.; Chong, J.; Burrows, C.J.; Wang, D. RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair–π and CH–π interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 9338–9348. [Google Scholar] [CrossRef]
- Chokkalla, A.K.; Mehta, S.L.; Vemuganti, R. Epitranscriptomic Modifications Modulate Normal and Pathological Functions in CNS. Transl. Stroke Res. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Anitha, A.; Nakamura, K.; Thanseem, I.; Matsuzaki, H.; Miyachi, T.; Tsujii, M.; Iwata, Y.; Suzuki, K.; Sugiyama, T.; Mori, N. Downregulation of the expression of mitochondrial electron transport complex genes in autism brains. Brain Pathol. 2013, 23, 294–302. [Google Scholar] [CrossRef]
- Gu, F.; Chauhan, V.; Kaur, K.; Brown, W.T.; LaFauci, G.; Wegiel, J.; Chauhan, A. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl. Psychiatry 2013, 3, e299. [Google Scholar] [CrossRef] [Green Version]
- Jones, A.; Thornton, C. Mitochondrial dynamics in the neonatal brain—A potential target following injury? Biosci. Rep. 2022, 42, BSR20211696. [Google Scholar] [CrossRef] [PubMed]
- Pecorelli, A.; Ferrara, F.; Messano, N.; Cordone, V.; Schiavone, M.L.; Cervellati, F.; Woodby, B.; Cervellati, C.; Hayek, J.; Valacchi, G. Alterations of mitochondrial bioenergetics, dynamics, and morphology support the theory of oxidative damage involvement in autism spectrum disorder. FASEB J. 2020, 34, 6521–6538. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, M.; Salazar, C.; Tiznado, W.; Ruiz, L.M. Alterations of Mitochondrial Biology in the Oral Mucosa of Chilean Children with Autism Spectrum Disorder (ASD). Cells 2019, 8, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Shi, X.; Boopathy, S.; McDonald, J.; Smith, A.W.; Chao, L.H. Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. Elife 2020, 9, e50973. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.M.; Rüegg, M.; Niemann, A.; Suter, U. Targeting and function of the mitochondrial fission factor GDAP1 are dependent on its tail-anchor. PLoS ONE 2009, 4, e5160. [Google Scholar] [CrossRef] [Green Version]
- Mao, K.; Klionsky, D.J. Participation of mitochondrial fission during mitophagy. Cell Cycle 2013, 12, 3131–3132. [Google Scholar] [CrossRef] [Green Version]
- Crespi, B.; Read, S.; Ly, A.; Hurd, P. AMBRA1, Autophagy, and the Extreme Male Brain Theory of Autism. Autism Res. Treat. 2019, 2019, 1968580. [Google Scholar] [CrossRef] [Green Version]
- Dalla Vecchia, E.; Mortimer, N.; Palladino, V.S.; Kittel-Schneider, S.; Lesch, K.P.; Reif, A.; Schenck, A.; Norton, W.H.J. Cross-species models of attention-deficit/hyperactivity disorder and autism spectrum disorder: Lessons from CNTNAP2, ADGRL3, and PARK2. Psychiatr. Genet. 2019, 29, 1–17. [Google Scholar] [CrossRef]
- Napoli, E.; Song, G.; Panoutsopoulos, A.; Riyadh, M.A.; Kaushik, G.; Halmai, J.; Levenson, R.; Zarbalis, K.S.; Giulivi, C. Beyond autophagy: A novel role for autism-linked Wdfy3 in brain mitophagy. Sci. Rep. 2018, 8, 11348. [Google Scholar] [CrossRef] [Green Version]
- Le Duc, D.; Giulivi, C.; Hiatt, S.M.; Napoli, E.; Panoutsopoulos, A.; Harlan De Crescenzo, A.; Kotzaeridou, U.; Syrbe, S.; Anagnostou, E.; Azage, M.; et al. Pathogenic WDFY3 variants cause neurodevelopmental disorders and opposing effects on brain size. Brain 2019, 142, 2617–2630. [Google Scholar] [CrossRef]
- Kirsten, T.B.; Chaves-Kirsten, G.P.; Chaible, L.M.; Silva, A.C.; Martins, D.O.; Britto, L.R.; Dagli, M.L.; Torrao, A.S.; Palermo-Neto, J.; Bernardi, M.M. Hypoactivity of the central dopaminergic system and autistic-like behavior induced by a single early prenatal exposure to lipopolysaccharide. J. Neurosci. Res. 2012, 90, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Kalivendi, S.; Zhang, H.; Joseph, J.; Nithipatikom, K.; Vásquez-Vivar, J.; Kalyanaraman, B. Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: Potential implications in intracellular fluorescence detection of superoxide. Free Radic. Biol. Med. 2003, 34, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Hwang, E.S. Fluorescence-based detection and quantification of features of cellular senescence. Methods Cell Biol. 2011, 103, 149–188. [Google Scholar] [CrossRef]
- Wojtala, A.; Bonora, M.; Malinska, D.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Methods to monitor ROS production by fluorescence microscopy and fluorometry. Methods Enzym. 2014, 542, 243–262. [Google Scholar] [CrossRef]
- Shpungin, S.; Dotan, I.; Abo, A.; Pick, E. Activation of the superoxide forming NADPH oxidase in a cell-free system by sodium dodecyl sulfate: Absolute lipid dependence of the solubilized enzyme. J. Biol. Chem. 1989, 264, 9195–9203. [Google Scholar] [CrossRef]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cieślik, M.; Zawadzka, A.; Czapski, G.A.; Wilkaniec, A.; Adamczyk, A. Developmental Stage-Dependent Changes in Mitochondrial Function in the Brain of Offspring Following Prenatal Maternal Immune Activation. Int. J. Mol. Sci. 2023, 24, 7243. https://doi.org/10.3390/ijms24087243
Cieślik M, Zawadzka A, Czapski GA, Wilkaniec A, Adamczyk A. Developmental Stage-Dependent Changes in Mitochondrial Function in the Brain of Offspring Following Prenatal Maternal Immune Activation. International Journal of Molecular Sciences. 2023; 24(8):7243. https://doi.org/10.3390/ijms24087243
Chicago/Turabian StyleCieślik, Magdalena, Aleksandra Zawadzka, Grzegorz A. Czapski, Anna Wilkaniec, and Agata Adamczyk. 2023. "Developmental Stage-Dependent Changes in Mitochondrial Function in the Brain of Offspring Following Prenatal Maternal Immune Activation" International Journal of Molecular Sciences 24, no. 8: 7243. https://doi.org/10.3390/ijms24087243
APA StyleCieślik, M., Zawadzka, A., Czapski, G. A., Wilkaniec, A., & Adamczyk, A. (2023). Developmental Stage-Dependent Changes in Mitochondrial Function in the Brain of Offspring Following Prenatal Maternal Immune Activation. International Journal of Molecular Sciences, 24(8), 7243. https://doi.org/10.3390/ijms24087243