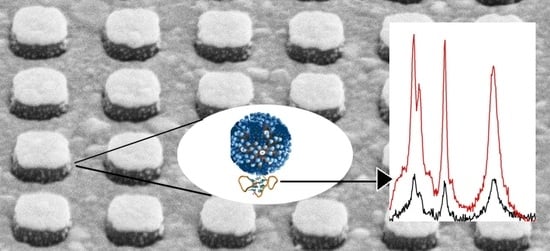
Multiplex Lithographic SERS Aptasensor for Detection of Several Respiratory Viruses in One Pot
Abstract
:1. Introduction
- −
- high sensitivity (up to 10−12–10−16 M of the substance in the sample);
- −
- the peaks of the SERS spectra have a narrow spectral band compared to fluorescence, which simplifies spectral separation in complex media;
- −
- the compatibility with antibodies, aptamers, and DNA probes allows specific detection of various proteins, genomes, viruses, cells, etc.
2. Results and Discussion
2.1. Specificity of the Aptamers
2.2. Setup of the Aptasensor
2.3. Multiplex Sensor
2.4. Comparison with Other Test Systems
3. Materials and Methods
3.1. Reagents
3.2. Lithographic SERS-Substrate
3.3. Viruses
3.4. Determination of Viral Particles in the Sample
3.5. Assembly of Aptamers
3.6. Biolayer Interferometry
3.7. Aptasensor Maintenance
3.8. SERS Measurements
3.9. Study of Surface Topology
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 5 April 2023).
- Aslan, A.; Aslan, C.; Zolbanin, N.M.; Jafari, R. Acute respiratory distress syndrome in COVID-19: Possible mechanisms and therapeutic management. Pneumonia 2021, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Saed, Y.A.; Song, W.; Wang, M.; Li, Y. Prevalence of non-SARS-CoV-2 respiratory pathogens and co-infection with SARS-CoV-2 in the early stage of COVID-19 epidemic. Pathogens 2022, 11, 1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Sebba, D.; Lastovich, A.G.; Kuroda, M.; Fallows, E.; Johnson, J.; Ahouidi, A.; Honko, A.N.; Fu, H.; Nielson, R.; Carruthers, E.; et al. A point-of-care diagnostic for differentiating Ebola from endemic febrile diseases. Sci. Transl. Med. 2018, 10, eaat0944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Huang, L.; Liu, B.; Ge, Q.; Dong, J.; Zhao, X. Rapid and ultrasensitive quantification of multiplex respiratory tract infection pathogen via lateral flow microarray based on SERS nanotags. Theranostics 2019, 9, 4849–4859. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, C.; Zheng, S.; Yang, X.; Han, H.; Dai, Y.; Xiao, R. Simultaneously ultrasensitive and quantitative detection of influenza A virus, SARS-CoV-2, and respiratory syncytial virus via multichannel magnetic SERS-based lateral flow immunoassay. Nanomedicine 2023, 47, 102624. [Google Scholar] [CrossRef]
- Kukushkin, V.; Ambartsumyan, O.; Astrakhantseva, A.; Gushchin, V.; Nikonova, A.; Dorofeeva, A.; Zverev, V.; Gambaryan, A.; Tikhonova, D.; Sovetnikov, T.; et al. Lithographic SERS aptasensor for ultrasensitive detection of SARS-CoV-2 in biological fluids. Nanomaterials 2022, 12, 3854. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, I.; Akitomi, J.; Boltz, D.A.; Horii, K.; Furuichi, M.; Waga, I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bizyaeva, A.A.; Bunin, D.A.; Moiseenko, V.L.; Gambaryan, A.S.; Balk, S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. The functional role of loops and flanking sequences of G-quadruplex aptamer to the hemagglutinin of influenza A virus. Int. J. Mol. Sci. 2021, 22, 2409. [Google Scholar] [CrossRef]
- Zhdanov, G.; Nyhrikova, E.; Meshcheryakova, N.; Kristavchuk, O.; Akhmetova, A.; Andreev, E.; Rudakova, E.; Gambaryan, A.; Yaminsky, I.; Aralov, A.; et al. A combination of membrane filtration and Raman-active DNA ligand greatly enhances sensitivity of SERS-based aptasensors for influenza A virus. Front. Chem. 2022, 10, 937180. [Google Scholar] [CrossRef]
- Percze, K.; Szakács, Z.; Scholz, É.; András, J.; Szeitner, Z.; van den Kieboom, C.H.; Ferwerda, G.; de Jonge, M.I.; Gyurcsányi, R.E.; Mészáros, T. Aptamers for respiratory syncytial virus detection. Sci. Rep. 2017, 7, 42794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Ambartsumyan, O.; Zhdanov, G.; Gribanyov, D.; Gushchin, V.; Tkachuk, A.; Rudakova, E.; Nikiforova, M.; Kuznetsova, N.; Popova, L.; et al. SERS-based aptasensor for rapid quantitative detection of SARS-CoV-2. Nanomaterials 2021, 11, 1394. [Google Scholar] [CrossRef]
- Ogorzaly, L.; Jourdan, S.; Mulholland, C. DNA Aptamers Specific of Adenovirus Types. Patent WO2020126435, 25 June 2020. [Google Scholar]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36 (Suppl. 2), W70–W74. [Google Scholar] [CrossRef] [Green Version]
- Novoseltseva, A.A.; Ivanov, N.M.; Novikov, R.A.; Tkachev, Y.V.; Bunin, D.A.; Gambaryan, A.S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. Structural and functional aspects of G-quadruplex aptamers which bind a broad range of influenza A viruses. Biomolecules 2020, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Kukushkin, V.I.; Grishina, Y.V.; Egorov, S.V.; Solov’ev, V.V.; Kukushkin, I.V. Combined dielectric and plasmon resonance for giant enhancement of Raman scattering. JEPT Lett. 2016, 103, 508–512. [Google Scholar] [CrossRef]
- Fedotova, Y.V.; Kukushkin, V.I.; Solovyev, V.V.; Kukushkin, I.V. Spoof plasmons enable giant Raman scattering enhancement in near-infrared region. Opt. Express 2019, 27, 32578–32586. [Google Scholar] [CrossRef]
- Wyllie, A.L. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Klimstra, W.B.; Tilston-Lunel, N.L.; Nambulli, S.; Boslett, J.; McMillen, C.M.; Gilliland, T.; Dunn, M.D.; Sun, C.; Wheeler, S.E.; Wells, A.; et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020, 101, 1156–1169. [Google Scholar] [CrossRef] [PubMed]
- Kramberger, P.; Ciringer, M.; Štrancar, A.; Peterka, M. Evaluation of nanoparticle tracking analysis for total virus particle determination. Virol. J. 2012, 9, 265. [Google Scholar] [CrossRef] [PubMed]
- McCormick, W.; Mermel, L.A. The basic reproductive number and particle-to-plaque ratio: Comparison of these two parameters of viral infectivity. Virol. J. 2021, 18, 92. [Google Scholar] [CrossRef]
- Duffy, A.M.; O’Doherty, A.M.; O’Brien, T.; Strappe, P.M. Purification of adenovirus and adeno-associated virus: Comparison of novel membrane-based technology to conventional techniques. Gene Ther. 2005, 12, S62–S72. [Google Scholar] [CrossRef] [Green Version]
- Yeh, Y.-T.; Gulino, K.; Zhang, Y.H.; Sabestien, A.; Chou, T.-W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V.; et al. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Nat. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef] [Green Version]
- Shanmukh, S.; Jones, L.; Driskell, J.; Zhao, Y.; Dluhy, R.; Tripp, R.A. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006, 6, 2630–2636. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Chen, L.; Liang, C.; Wei, Q.; Chen, Y.; Wang, J. Porous carbon films decorated with silver nanoparticles as a sensitive SERS substrate, and their application to virus identification. Microchim. Acta 2017, 184, 3505–3511. [Google Scholar] [CrossRef]
- Kukushkin, V.; Kristavchuk, O.; Andreev, E.; Meshcheryakova, N.; Zaborova, O.; Gambaryan, A.; Nechaev, A.; Zavyalova, E. Aptamer-coated track-etched membranes with a nanostructured silver layer for single virus detection in biological fluids. Front. Bioeng. Biotechnol. 2023, 10, 1076749. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Park, S.K.; Joung, Y.; Kang, T.; Lee, M.K.; Choo, J. SERS-based dual-mode DNA aptasensors for rapid classification of SARS-CoV-2 and influenza A/H1N1 infection. Sens. Actuators B Chem. 2022, 355, 131324. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Yi, S.Y.; Hwang, A.; Eom, G.; Sim, J.; Jeong, J.; Lim, E.-K.; Chung, B.H.; Kim, B.; Jung, J.; et al. Facile and sensitive detection of influenza viruses using SERS antibody probes. RSC Adv. 2016, 6, 84415–84419. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-J.; Wu, C.-Y.; Li, T.; Hsiao, P.-W.; Chang, D.-K. A rapid and sensitive early diagnosis of influenza virus subtype via surface enhanced Raman scattering. J. Biosens. Bioelectron. 2014, 5, 2. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, B.C.; Oh, B.K.; Choi, J.W. Rapid and sensitive determination of HIV-1 virus based on surface enhanced Raman spectroscopy. J. Biomed. Nanotechnol. 2015, 11, 2223–2230. [Google Scholar] [CrossRef]
- Herrmann, B.; Larsson, C.; Zweygberg, B.W. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription-PCR: Comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA). J. Clin. Microbiol. 2001, 39, 134–138. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.Y.; Jian, M.J.; Chang, C.K.; Lin, J.C.; Yeh, K.M.; Chen, C.W.; Chiu, S.K.; Wang, Y.H.; Liao, S.J.; Li, S.Y.; et al. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg. Microbes Infect. 2021, 10, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Yoon, S.Y.; Lim, C.S.; Yoon, J. Comparison of three molecular diagnostic assays for SARS-CoV-2 detection: Evaluation of analytical sensitivity and clinical performance. Clin. Lab. Anal. 2022, 36, e24242. [Google Scholar] [CrossRef]
- Yang, J.; Han, Y.; Zhang, R.; Zhang, R.; Li, J. Comparison of analytical sensitivity of SARS-CoV-2 molecular detection kits. Int. J. Infect. Dis. 2021, 111, 233–241. [Google Scholar] [CrossRef]
- Luo, S.; Xie, Z.; Xie, L.; Liu, J.; Xie, Z.; Deng, X.; Huang, L.; Huang, J.; Zeng, T.; Khan, M.I. Reverse-transcription, loop-mediated isothermal amplification assay for the sensitive and rapid detection of H10 subtype avian influenza viruses. Virol. J. 2015, 12, 145. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y.; et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.-H.; To, K.K.W.; Chan, J.F.W.; Li, C.P.Y.; Chen, H.; Yuen, K.-Y. Analytical sensitivity of seven point-of-care influenza virus detection tests and two molecular tests for detection of avian origin H7N9 and swine origin H3N2 variant influenza A viruses. J. Clin. Microbiol. 2013, 51, 3160–3161. [Google Scholar] [CrossRef] [Green Version]
- Instructions for Use of QuickVue SARS Antigen Test. 2021. Available online: https://www.fda.gov/media/144668/download (accessed on 25 December 2022).
- Masson, L.; Mazza, A.; De Crescenzo, G. Determination of affinity and kinetic rate constants using surface plasmon resonance. Methods Mol. Biol. 2000, 145, 189–201. [Google Scholar] [CrossRef] [PubMed]







| Aptamer | Influenza A Virus | ADV 5 | RSV | SARS-CoV-2 |
|---|---|---|---|---|
| RHA0385 | (2.1 ± 0.3) × 10−14 | Low binding | Low binding | Low binding |
| ADV-20 | (2.6 ± 0.2) × 10−12 | (1.1 ± 0.2) × 10−14 | (3.8 ± 0.5) × 10−13 | (5 ± 2) × 10−11 |
| H8 | (3.5 ± 1.1) × 10−12 | Low binding | (3.1 ± 0.8) × 10−14 | Low binding |
| RBD-1C | Low binding | Low binding | Low binding | (7.1 ± 0.6) × 10−14 |
| # of the Sensor | Spot | Detectable Virus in the Spot | Virus Content, VP/mL | Analytical Signal from Labeled Aptamers | |||
|---|---|---|---|---|---|---|---|
| SERS, a.u. | SEL, a.u. | SERS/SEL | Result | ||||
| 1 | a | Adenovirus type 5 | 0 | 11,800 ± 500 | 29,100 ± 700 | 0.41 ± 0.01 | ≤0.44 → neg. |
| b | Influenza A H7N1 | 0 | 5300 ± 200 | 18,800 ± 600 | 0.28 ± 0.01 | ≥0.25 → neg. | |
| c | SARS-CoV-2 | 1 × 105 | 2800 ± 100 | 9200 ± 300 | 0.31 ± 0.01 | <0.32 → pos. | |
| d | RSV A2 | 3 × 106 | 2600 ± 100 | 12,200 ± 300 | 0.21 ± 0.01 | <0.31 → pos. | |
| 2 | a | Adenovirus type 5 | 7 × 105 | 10,000 ± 400 | 17,900 ± 700 | 0.56 ± 0.01 | >0.44 → pos. |
| b | Influenza A H7N1 | 6 × 106 | 3900 ± 100 | 19,500 ± 300 | 0.20 ± 0.01 | <0.25 → pos. | |
| c | SARS-CoV-2 | 0 | 2700 ± 100 | 8200 ± 200 | 0.33 ± 0.01 | ≥0.32 → neg. | |
| d | RSV A2 | 0 | 2500 ± 100 | 7800 ± 500 | 0.32 ± 0.01 | ≥0.31 → neg. | |
| 3 | a | Adenovirus type 5 | 0 | 10,000 ± 400 | 23,300 ± 800 | 0.43 ± 0.01 | ≤0.44 → neg. |
| b | Influenza A H7N1 | 6 × 106 | 5100 ± 100 | 2300 ± 300 | 0.22 ± 0.01 | <0.25 → pos. | |
| c | SARS-CoV-2 | 1 × 104 | 2900 ± 200 | 12,500 ± 300 | 0.24 ± 0.01 | <0.32 → pos. | |
| d | RSV A2 | 3 × 105 | 2600 ± 100 | 8500 ± 200 | 0.30 ± 0.01 | <0.31 → pos. | |
| 4 | a | Adenovirus type 5 | 7 × 104 | 8800 ± 400 | 15,800 ± 500 | 0.56 ± 0.01 | >0.44 → pos. |
| b | Influenza A H7N1 | 6 × 105 | 3500 ± 100 | 15,200 ± 100 | 0.23 ± 0.01 | <0.25 → pos. | |
| c | SARS-CoV-2 | 1 × 103 | 3400 ± 200 | 10,800 ± 200 | 0.31 ± 0.01 | <0.32 → pos. | |
| d | RSV A2 | 0 | 2300 ± 100 | 6800 ± 200 | 0.33 ± 0.01 | ≥0.31 → neg. | |
| 5 | a | Adenovirus type 5 | 0 | 8200 ± 300 | 26,200 ± 900 | 0.31 ± 0.01 | ≤0.44 → neg. |
| b | Influenza A H7N1 | 6 × 106 | 3700 ± 100 | 18,700 ± 100 | 0.20 ± 0.01 | <0.25 → pos. | |
| c | SARS-CoV-2 | 1 × 105 | 2900 ± 100 | 11,300 ± 500 | 0.26 ± 0.01 | <0.32 → pos. | |
| d | RSV A2 | 0 | 2400 ± 100 | 7000 ± 200 | 0.33 ± 0.01 | ≥0.31 → neg. | |
| 6 | a | Adenovirus type 5 | 7 × 105 | 11,700 ± 300 | 21,000 ± 900 | 0.56 ± 0.01 | >0.44 → pos. |
| b | Influenza A H7N1 | 0 | 4900 ± 100 | 16,700 ± 100 | 0.29 ± 0.01 | ≥0.25 → neg. | |
| c | SARS-CoV-2 | 0 | 2800 ± 200 | 8200 ± 300 | 0.34 ± 0.01 | ≥0.32 → neg. | |
| d | RSV A2 | 3 × 106 | 2400 ± 100 | 8300 ± 100 | 0.29 ± 0.01 | <0.31 → pos. | |
| Analytical Technique | Virus Type | Analytical Performance | Ref. | ||||
|---|---|---|---|---|---|---|---|
| Limit of Detection | Quantification | Time of Analysis | Multiplex Analysis | ||||
| Observed | Recalculated | ||||||
| SERS without recognizing elements | Rheovirus, rinovirus, influenza A, parainfluenza | 102 EID50/mL | 104 VP/mL | No | 15 min | Not reported | [25] |
| Respiratory syncytial virus | 100 pfu/mL | 3 × 105 VP/mL | Yes | 1 h | Not reported | [26] | |
| Circovirus, parvovirus, pseudorabies | 1 × 107 VP/mL | 1 × 107 VP/mL | No | 15 min | Not reported | [27] | |
| Aptamer-based SERS | Influenza A | 2.2 × 10−5 HAU/mL | 103 VP/mL | No | 15 min | Not reported | [11] |
| 10 VP/mL | 10 VP/mL | Yes | 15 min | Not reported | [28] | ||
| Influenza A/ SARS-CoV-2 | 1.06 HAU/mL 0.95 pfu/mL | 5 × 107 VP/mL 7 × 105 VP/mL | Yes | 15 min | Yes | [29] | |
| SARS-CoV-2 | 100 copies/mL | 100 VP/mL | Yes | 15 min | Not reported | [8] | |
| Influenza A/ SARS-CoV-2/ RSV/ Adenovirus | 600 VP/mL 100 VP/mL 3 × 103 VP/mL 70 VP/mL | 600 VP/mL 100 VP/mL 3 × 103 VP/mL 70 VP/mL | No | 17 min | Yes | This work | |
| Antibody-based SERS | Influenza A | 4 × 103 TCID50/mL | 4 × 105 VP/mL | Yes | 3.5 h | Not reported | [30] |
| 30 ng/mL | 1.8 × 108 VP/mL | Yes | 2 h | Not reported | [31] | ||
| Influenza A/ Adenovirus | 50 pfu/mL 10 pfu/mL | 3 × 107 VP/mL 500 VP/mL | Yes | 30 min | Yes | [4] | |
| Ebola/Lassa viruses | 105 pfu/mL | 106 VP/mL | Yes | 30 min | Yes | [5] | |
| Influenza A/ SARS-CoV-2/ RSV | 85 copies/mL 8 pg/mL 8 pg/mL | 85 VP/mL 5 × 104 VP/mL 5 × 104 VP/mL | Yes | 30 min | Yes | [7] | |
| Human immunodeficiency virus | 35 pg/mL | 2 × 105 VP/mL | Yes | 12 h | Not reported | [32] | |
| DNA probe-based SERS | Influenza A and B/parainfluenza/RSV/parainfluenza/adenovirus | 30–41 fM | 2 × 107 VP/mL | Yes | 10 min | Yes | [6] |
| RT PCR | Influenza A | 3 × 102–1.2 × 103 VP/mL | 3 × 102–1.2 × 103 VP/mL | Yes | 2–3 h | Yes | [33,34] |
| SARS-CoV-2 | 102–1.2 × 103 VP/mL | 102–1.2 × 103 VP/mL | Yes | 2–3 h | [34,35,36] | ||
| RT LAMP | Influenza A | 104 VP/mL | 104 VP/mL | Yes | 1 h | Yes | [37] |
| SARS-CoV-2 | 2 × 104 VP/mL | 2 × 104 VP/mL | Yes | 1 h | [38] | ||
| Antibody-based test strip | Influenza A | 20 TCID50/mL | 1 × 106 VP/mL | No | 10–15 min | Yes | [39] |
| SARS-CoV-2 | 7.6 × 103 TCID50/mL | 5 × 108 VP/mL | No | 15 min | [40] | ||
| Code | Sequence (5→3) | Target |
|---|---|---|
| Biotin-RHA0386 | (Biotin)-TTGGGGTTATTTTGGGAGGGCGGGGGTT | Hemagglutinin of influenza A |
| SH-RHA0386-Cy3 | (SH-(CH2)6)-TTGGGGTTATTTTGGGAGGGCGGGGGTT-(Cy3) | Hemagglutinin of influenza A |
| Biotin-RBD-1C | (Biotin)-CAGCACCGACCTTGTGCTTTGGGAG TGCTGGTCCAAGGGCGTTAATGGACA | S protein of SARS-CoV-2 |
| SH-RBD-1C-Cy3 | (SH-(CH2)6)-CAGCACCGACCTTGTGCTTTGGGAG TGCTGGTCCAAGGGCGTTAATGGACA-(Cy3) | S protein of SARS-CoV-2 |
| Biotin-H8 | (Biotin)-AGTGCGGTGAGCCGTCGGACATACAAATAC | RSV A2 virions |
| SH-H8-TAMRA | (SH-(CH2)6)-AGTGCGGTGAGCCGTCGGACATACAA ATAC-(TAMRA) | RSV A2 virions |
| Biotin-ADV-20 | (Biotin)-GTGCCAGCTATGCCATTGGCGGGTCGTCC AATTCGAGAGGTCCCCTAGCGTCTATCTCTGCTGC | Adenovirus type 2 |
| SH-ADV-20-Cy3 | (SH-(CH2)6)-GTGCCAGCTATGCCATTGGCGGGTCGT CCAATTCGAGAGGTCCCCTAGCGTCTATCTCTGCTGC-(Cy3) | Adenovirus type 2 |
| Virus | Number of Virions per mL |
|---|---|
| Influenza A virus | 6.1 × 109 |
| SARS-CoV-2 | 1.0 × 108 * |
| Respiratory syncytial virus | 3.1 × 109 |
| Adenovirus 5 | 7.4 × 108 |
| Influenza B virus | 6.0 × 109 |
| Newcastle disease virus | 3.6 × 1010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kukushkin, V.; Ambartsumyan, O.; Subekin, A.; Astrakhantseva, A.; Gushchin, V.; Nikonova, A.; Dorofeeva, A.; Zverev, V.; Keshek, A.; Meshcheryakova, N.; et al. Multiplex Lithographic SERS Aptasensor for Detection of Several Respiratory Viruses in One Pot. Int. J. Mol. Sci. 2023, 24, 8081. https://doi.org/10.3390/ijms24098081
Kukushkin V, Ambartsumyan O, Subekin A, Astrakhantseva A, Gushchin V, Nikonova A, Dorofeeva A, Zverev V, Keshek A, Meshcheryakova N, et al. Multiplex Lithographic SERS Aptasensor for Detection of Several Respiratory Viruses in One Pot. International Journal of Molecular Sciences. 2023; 24(9):8081. https://doi.org/10.3390/ijms24098081
Chicago/Turabian StyleKukushkin, Vladimir, Oganes Ambartsumyan, Alexei Subekin, Anna Astrakhantseva, Vladimir Gushchin, Alexandra Nikonova, Anastasia Dorofeeva, Vitaly Zverev, Anna Keshek, Nadezda Meshcheryakova, and et al. 2023. "Multiplex Lithographic SERS Aptasensor for Detection of Several Respiratory Viruses in One Pot" International Journal of Molecular Sciences 24, no. 9: 8081. https://doi.org/10.3390/ijms24098081
APA StyleKukushkin, V., Ambartsumyan, O., Subekin, A., Astrakhantseva, A., Gushchin, V., Nikonova, A., Dorofeeva, A., Zverev, V., Keshek, A., Meshcheryakova, N., Zaborova, O., Gambaryan, A., & Zavyalova, E. (2023). Multiplex Lithographic SERS Aptasensor for Detection of Several Respiratory Viruses in One Pot. International Journal of Molecular Sciences, 24(9), 8081. https://doi.org/10.3390/ijms24098081