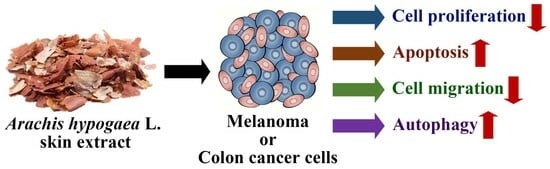
Inhibition of Autophagy Aggravates Arachis hypogaea L. Skin Extracts-Induced Apoptosis in Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Differential Cytotoxic Activities of Peanut Skin Fractions against Melanoma and CRC Cells
2.2. Antiproliferative Effects of Peanut Skin Extracts on Melanoma and CRC Cells
2.3. Melanoma and CRC Cells Undergo Apoptosis in Response to AHE and AHE-2
2.4. Peanut Skin Extracts Significantly Inhibit Cell Migration in Melanoma and CRC Cells
2.5. Inhibition of Autophagy Enhances AHE- and AHE-2-Induced Cytotoxicity and Apoptosis in Melanoma and CRC Cells
2.6. The Chemoprofiling of AHE-2
3. Discussion
4. Materials and Methods
4.1. Preparation of Peanut Skin Fractions
4.2. Cell culture and Antibodies
4.3. Cell Viability Assay
4.4. Colony Formation Assay
4.5. Cell Cycle Analysis
4.6. Annexin V-FITC/PI Apoptosis Analysis
4.7. Western Blot Analysis
4.8. Scratch Wound-Healing Assay
4.9. Transwell Migration Assay
4.10. Transient Transfection and GFP-LC3 Fluorescence Imaging Analysis
4.11. HPLC Analysis
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; the International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Mann, J. Natural products in cancer chemotherapy: Past, present and future. Nat. Rev. Cancer 2002, 2, 143–148. [Google Scholar] [CrossRef]
- Lachance, J.C.; Radhakrishnan, S.; Madiwale, G.; Guerrier, S.; Vanamala, J.K.P. Targeting hallmarks of cancer with a food-system-based approach. Nutrition 2020, 69, 110563. [Google Scholar] [CrossRef]
- Lefranc, F.; Tabanca, N.; Kiss, R. Assessing the anticancer effects associated with food products and/or nutraceuticals using in vitro and in vivo preclinical development-related pharmacological tests. Semin. Cancer Biol. 2017, 46, 14–32. [Google Scholar] [CrossRef]
- Ojiewo, C.O.; Janila, P.; Bhatnagar-Mathur, P.; Pandey, M.K.; Desmae, H.; Okori, P.; Mwololo, J.; Ajeigbe, H.; Njuguna-Mungai, E.; Muricho, G.; et al. Advances in Crop Improvement and Delivery Research for Nutritional Quality and Health Benefits of Groundnut (Arachis hypogaea L.). Front. Plant Sci. 2020, 11, 29. [Google Scholar] [CrossRef]
- Kim, M.Y.; Kim, H.J.; Lee, Y.Y.; Kim, M.H.; Lee, J.Y.; Kang, M.S.; Koo, B.C.; Lee, B.W. Antioxidant and anti-inflammatory effects of Peanut (Arachishypogaea L.) skin extracts of various cultivars in oxidative-damaged HepG2 cells and LPS-induced raw 264.7 macrophages. Food Sci. Nutr. 2021, 9, 973–984. [Google Scholar] [CrossRef]
- Kyei, S.K.; Eke, W.I.; Abdul-Karim, H.; Darko, G.; Akaranta, O. Phytochemicals from peanut (Arachis hypogaea L.) skin extract with potential for pharmacological activity. Curr. Bioact. Compd. 2021, 17, 38–56. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Comito, F.; Pagani, R.; Grilli, G.; Sperandi, F.; Ardizzoni, A.; Melotti, B. Emerging Novel Therapeutic Approaches for Treatment of Advanced Cutaneous Melanoma. Cancers 2022, 14, 271. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Lombardi, L.; Morelli, F.; Cinieri, S.; Santini, D.; Silvestris, N.; Fazio, N.; Orlando, L.; Tonini, G.; Colucci, G.; Maiello, E. Adjuvant colon cancer chemotherapy: Where we are and where we’ll go. Cancer Treat. Rev. 2010, 36 (Suppl. S3), S34–S41. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Goldman, E.; Fisher, J.L. Discrepancies in cancer mortality estimates. Arch. Med. Res. 2006, 37, 548–551. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Lim, S.M.; Mohamad Hanif, E.A.; Chin, S.F. Is targeting autophagy mechanism in cancer a good approach? The possible double-edge sword effect. Cell Biosci. 2021, 11, 56. [Google Scholar] [CrossRef]
- Alharbi, Y.M.; Bima, A.I.; Elsamanoudy, A.Z. An Overview of the Perspective of Cellular Autophagy: Mechanism, Regulation, and the Role of Autophagy Dysregulation in the Pathogenesis of Diseases. J. Microsc. Ultrastruct. 2021, 9, 47–54. [Google Scholar] [CrossRef]
- Tan, Q.; Wang, M.; Yu, M.; Zhang, J.; Bristow, R.G.; Hill, R.P.; Tannock, I.F. Role of Autophagy as a Survival Mechanism for Hypoxic Cells in Tumors. Neoplasia 2016, 18, 347–355. [Google Scholar] [CrossRef]
- Altman, J.K.; Szilard, A.; Goussetis, D.J.; Sassano, A.; Colamonici, M.; Gounaris, E.; Frankfurt, O.; Giles, F.J.; Eklund, E.A.; Beauchamp, E.M.; et al. Autophagy is a survival mechanism of acute myelogenous leukemia precursors during dual mTORC2/mTORC1 targeting. Clin. Cancer Res. 2014, 20, 2400–2409. [Google Scholar] [CrossRef]
- Masui, A.; Hamada, M.; Kameyama, H.; Wakabayashi, K.; Takasu, A.; Imai, T.; Iwai, S.; Yura, Y. Autophagy as a Survival Mechanism for Squamous Cell Carcinoma Cells in Endonuclease G-Mediated Apoptosis. PLoS ONE 2016, 11, e0162786. [Google Scholar] [CrossRef]
- Chen, L.; Yan, F.; Chen, W.; Zhao, L.; Zhang, J.; Lu, Q.; Liu, R. Procyanidin from peanut skin induces antiproliferative effect in human prostate carcinoma cells DU145. Chem. Biol. Interact. 2018, 288, 12–23. [Google Scholar] [CrossRef]
- Galgut, J.M.; Ali, S.A. Effect and mechanism of action of resveratrol: A novel melanolytic compound from the peanut skin of Arachis hypogaea. J. Recept. Signal Transduct. Res. 2011, 31, 374–380. [Google Scholar] [CrossRef]
- Saenglee, S.; Senawong, G.; Jogloy, S.; Sripa, B.; Senawong, T. Peanut testa extracts possessing histone deacetylase inhibitory activity induce apoptosis in cholangiocarcinoma cells. Biomed. Pharmacother. 2018, 98, 233–241. [Google Scholar] [CrossRef]
- Topacio, B.R.; Zatulovskiy, E.; Cristea, S.; Xie, S.; Tambo, C.S.; Rubin, S.M.; Sage, J.; Koivomagi, M.; Skotheim, J.M. Cyclin D-Cdk4,6 Drives Cell-Cycle Progression via the Retinoblastoma Protein’s C-Terminal Helix. Mol. Cell 2019, 74, 758–770.e754. [Google Scholar] [CrossRef]
- Roomi, M.W.; Bhanap, B.; Niedzwiecki, A.; Rath, M. Progress of Tumor Growth and Metastasis After Inoculation of B16FO Melanoma Cells in Kidney of Female Nude Mice Is Inhibited by a Novel Nutrient Mixture. Integr. Cancer Ther. 2019, 18, 1534735419832365. [Google Scholar] [CrossRef]
- Pretzsch, E.; Bosch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M. Mechanisms of Metastasis in Colorectal Cancer and Metastatic Organotropism: Hematogenous versus Peritoneal Spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef]
- White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 2012, 12, 401–410. [Google Scholar] [CrossRef]
- Hu, Y.L.; Jahangiri, A.; Delay, M.; Aghi, M.K. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res. 2012, 72, 4294–4299. [Google Scholar] [CrossRef]
- Zou, Z.; Yuan, Z.; Zhang, Q.; Long, Z.; Chen, J.; Tang, Z.; Zhu, Y.; Chen, S.; Xu, J.; Yan, M.; et al. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy 2012, 8, 1798–1810. [Google Scholar] [CrossRef]
- Mohd Sairazi, N.S.; Sirajudeen, K.N.S. Natural Products and Their Bioactive Compounds: Neuroprotective Potentials against Neurodegenerative Diseases. Evid.-Based Complement. Altern. Med. 2020, 2020, 6565396. [Google Scholar] [CrossRef]
- Rajesh, E.; Sankari, L.S.; Malathi, L.; Krupaa, J.R. Naturally occurring products in cancer therapy. J. Pharm. Bioallied Sci. 2015, 7, S181–S183. [Google Scholar] [CrossRef]
- Yun, C.W.; Kim, H.J.; Lee, S.H. Therapeutic Application of Diverse Marine-derived Natural Products in Cancer Therapy. Anticancer. Res. 2019, 39, 5261–5284. [Google Scholar] [CrossRef]
- Tahergorabi, Z.; Abedini, M.R.; Mitra, M.; Fard, M.H.; Beydokhti, H. "Ziziphus jujuba": A red fruit with promising anticancer activities. Pharmacogn. Rev. 2015, 9, 99–106. [Google Scholar] [CrossRef]
- Wali, A.F.; Majid, S.; Rasool, S.; Shehada, S.B.; Abdulkareem, S.K.; Firdous, A.; Beigh, S.; Shakeel, S.; Mushtaq, S.; Akbar, I.; et al. Natural products against cancer: Review on phytochemicals from marine sources in preventing cancer. Saudi Pharm. J. 2019, 27, 767–777. [Google Scholar] [CrossRef]
- Amin, A.R.; Kucuk, O.; Khuri, F.R.; Shin, D.M. Perspectives for cancer prevention with natural compounds. J. Clin. Oncol. 2009, 27, 2712–2725. [Google Scholar] [CrossRef]
- Choudhari, A.S.; Mandave, P.C.; Deshpande, M.; Ranjekar, P.; Prakash, O. Phytochemicals in Cancer Treatment: From Preclinical Studies to Clinical Practice. Front. Pharmacol. 2019, 10, 1614. [Google Scholar] [CrossRef]
- Bailon-Moscoso, N.; Cevallos-Solorzano, G.; Romero-Benavides, J.C.; Orellana, M.I. Natural Compounds as Modulators of Cell Cycle Arrest: Application for Anticancer Chemotherapies. Curr. Genomics 2017, 18, 106–131. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef]
- Tchakarska, G.; Sola, B. The double dealing of cyclin D1. Cell Cycle 2020, 19, 163–178. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef]
- Tas, F. Metastatic behavior in melanoma: Timing, pattern, survival, and influencing factors. J. Oncol. 2012, 2012, 647684. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Liu, Y.; Zhang, C.; Li, H.; Lai, B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med. 2020, 9, 361–373. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal 2014, 20, 460–473. [Google Scholar] [CrossRef]
- Alvarez-Meythaler, J.G.; Garcia-Mayea, Y.; Mir, C.; Kondoh, H.; ME, L.L. Autophagy Takes Center Stage as a Possible Cancer Hallmark. Front. Oncol. 2020, 10, 586069. [Google Scholar] [CrossRef]
- Patergnani, S.; Missiroli, S.; Morciano, G.; Perrone, M.; Mantovani, C.M.; Anania, G.; Fiorica, F.; Pinton, P.; Giorgi, C. Understanding the Role of Autophagy in Cancer Formation and Progression Is a Real Opportunity to Treat and Cure Human Cancers. Cancers 2021, 13, 5622. [Google Scholar] [CrossRef]
- Rahmati, M.; Ebrahim, S.; Hashemi, S.; Motamedi, M.; Moosavi, M.A. New insights on the role of autophagy in the pathogenesis and treatment of melanoma. Mol. Biol. Rep. 2020, 47, 9021–9032. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Zhang, X.Y.; Wang, X.F.; Sun, B.C. Autophagy enhances the aggressiveness of human colorectal cancer cells and their ability to adapt to apoptotic stimulus. Cancer Biol. Med. 2012, 9, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yun, C.W.; Han, Y.S.; Kim, S.; Jeong, D.; Kwon, H.Y.; Kim, H.; Baek, M.J.; Lee, S.H. Melatonin and 5-fluorouracil co-suppress colon cancer stem cells by regulating cellular prion protein-Oct4 axis. J. Pineal Res. 2018, 65, e12519. [Google Scholar] [CrossRef] [PubMed]
- Towers, C.G.; Thorburn, A. Therapeutic Targeting of Autophagy. EBioMedicine 2016, 14, 15–23. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, C.-H.; Huang, H.-C.; Lin, K.-J.; Liu, J.-M.; Chen, G.-L.; Yeh, Y.-H.; Lu, T.-L.; Lin, H.-W.; Lu, M.-T.; Chu, P.-C. Inhibition of Autophagy Aggravates Arachis hypogaea L. Skin Extracts-Induced Apoptosis in Cancer Cells. Int. J. Mol. Sci. 2024, 25, 1345. https://doi.org/10.3390/ijms25021345
Tsai C-H, Huang H-C, Lin K-J, Liu J-M, Chen G-L, Yeh Y-H, Lu T-L, Lin H-W, Lu M-T, Chu P-C. Inhibition of Autophagy Aggravates Arachis hypogaea L. Skin Extracts-Induced Apoptosis in Cancer Cells. International Journal of Molecular Sciences. 2024; 25(2):1345. https://doi.org/10.3390/ijms25021345
Chicago/Turabian StyleTsai, Chia-Hung, Hui-Chi Huang, Kuan-Jung Lin, Jui-Ming Liu, Guan-Lin Chen, Yi-Hsien Yeh, Te-Ling Lu, Hsiang-Wen Lin, Meng-Tien Lu, and Po-Chen Chu. 2024. "Inhibition of Autophagy Aggravates Arachis hypogaea L. Skin Extracts-Induced Apoptosis in Cancer Cells" International Journal of Molecular Sciences 25, no. 2: 1345. https://doi.org/10.3390/ijms25021345
APA StyleTsai, C. -H., Huang, H. -C., Lin, K. -J., Liu, J. -M., Chen, G. -L., Yeh, Y. -H., Lu, T. -L., Lin, H. -W., Lu, M. -T., & Chu, P. -C. (2024). Inhibition of Autophagy Aggravates Arachis hypogaea L. Skin Extracts-Induced Apoptosis in Cancer Cells. International Journal of Molecular Sciences, 25(2), 1345. https://doi.org/10.3390/ijms25021345