Evidence for Plio-Pleistocene Duck Mussel Refugia in the Azov Sea River Basins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, PCR and Sequencing
2.2. Phylogenetic Analyses and Divergence Time Estimates
2.3. Demographic History and Molecular Dating Analysis
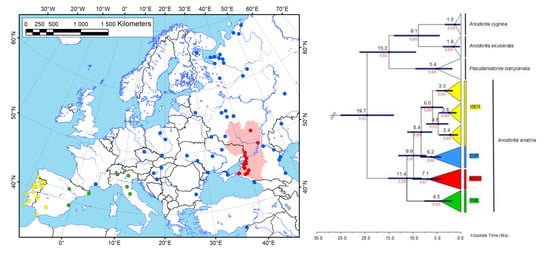
3. Results
3.1. Phylogenetic Reconstruction and Phylogeography
3.2. Historical Demography and Divergence Time Estimates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 454–464. [Google Scholar] [CrossRef]
- Froufe, E.; Lopes-Lima, M.; Riccardi, N.; Zaccara, S.; Vanetti, I.; Lajtner, J.; Teixeira, A.; Varandas, S.; Prié, V.; Zieritz, A.; et al. Lifting the curtain on the freshwater mussel diversity from the Italian Peninsula and Croatian Adriatic coast. Biodivers. Conserv. 2017, 26, 3255–3274. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, C.; Wolff, C.; Glöer, P.; Wilke, T. Concurrent evolution of ancient sister lakes and sister species: The freshwater gastropod genus Radix in lakes Ohrid and Prespa. Hydrobiologia 2008, 615, 157–167. [Google Scholar] [CrossRef]
- Grabowski, M.; Mamos, T.; Bącela-Spychalska, K.; Rewicz, T.; Wattier, R.A. Neogene paleogeography provides context for understanding the origin and spatial distribution of cryptic diversity in a widespread Balkan freshwater amphipod. PeerJ 2017, 5, e3016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krijgsman, W.; Tesakov, A.; Yanina, T.; Lazarev, S.; Danukalova, G.; Van Baak, C.G.C.; Agustí, J.; Alçiçek, M.C.; Aliyeva, E.; Bista, D.; et al. Quaternary time scales for the Pontocaspian domain: Interbasinal connectivity and faunal evolution. Earth Sci. Rev. 2019, 188, 1–40. [Google Scholar] [CrossRef]
- Van Baak, C.G.C.; Mandic, O.; Lazar, I.; Stoica, M.; Krijgsman, W. The Slanicul de Buzau section, a unit stratotype for the Romanian stage of the Dacian Basin (Plio-Pleistocene, Eastern Paratethys). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2015, 440, 594–613. [Google Scholar] [CrossRef]
- Jorissen, E.L.; de Leeuw, A.; van Baak, C.G.C.; Mandic, O.; Stoica, M.; Abels, H.A.; Krijgsman, W. Sedimentary architecture and depositional controls of a Pliocene river-dominated delta in the semi-isolated Dacian Basin, Black Sea. Sediment. Geol. 2018, 368, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Borzenkova, I.I. Izmenenie Klimata v Kainozoe (Climatic Changes in the Cenozoic); Gidrometeoizdat: St.-Petersburg, Russia, 1992. [Google Scholar]
- Levin, B.A.; Gandlin, A.A.; Simonov, E.S.; Levina, M.A.; Barmintseva, A.E.; Japoshvili, B.; Mugue, N.S.; Mumladze, L.; Mustafayev, N.J.; Pashkov, A.N.; et al. Phylogeny, phylogeography and hybridization of Caucasian barbels of the genus Barbus (Actinopterygii, Cyprinidae). Mol. Phylogenet. Evol. 2019, 135, 31–44. [Google Scholar] [CrossRef]
- Nahavandi, N.; Ketmaier, V.; Plath, M.; Tiedemann, R. Diversification of Ponto-Caspian aquatic fauna: Morphology and molecules retrieve congruent evolutionary relationships in Pontogammarus maeoticus (Amphipoda: Pontogammaridae). Mol. Phylogenet. Evol. 2013, 69, 1063–1076. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Kondakov, A.V.; Konopleva, E.S.; Vikhrev, I.V.; Aksenova, O.V.; Aksenov, A.S.; Bespalaya, Y.V.; Borovskoy, A.V.; Danilov, P.P.; Dvoryankin, G.A.; et al. Integrative taxonomy, biogeography and conservation of freshwater mussels (Unionidae) in Russia. Sci. Rep. 2020, 10, 3072. [Google Scholar] [CrossRef] [Green Version]
- Soroka, M. Identification of gender-associated mitochondrial haplotypes in Anodonta anatina (Bivalvia: Unionidae). Folia Malacol. 2008, 16, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Froufe, E.; Sobral, C.; Teixeira, A.; Sousa, R.; Varandas, S.; Aldridge, D.; Lopes-Lima, M. Genetic diversity of the pan-European freshwater mussel Anodonta anatina (Bivalvia: Unionoida) based on CO1: New phylogenetic insights and implications for conservation. Aquat. Conserv. Mar. Freshw. Ecosyst. 2014, 24, 561–574. [Google Scholar] [CrossRef] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Bolotov, I.N.; Kondakov, A.V.; Vikhrev, I.V.; Aksenova, O.V.; Bespalaya, Y.V.; Gofarov, M.Y.; Kolosova, Y.S.; Konopleva, E.S.; Spitsyn, V.M.; Tanmuangpak, K.; et al. Ancient river inference explains exceptional Oriental freshwater mussel radiations. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tomilova, A.A.; Kondakov, A.V.; Kisil, O.Y. Usage of transcribed spacers ITS1 and ITS2 for identification of freshwater mussels of the genera Anodonta and Pseudanodonta (Bivalvia: Unionidae: Anodontinae). Zhurnal Obshchei Biologii 2019, 80, 364–371. [Google Scholar]
- Klishko, O.K.; Lopes-Lima, M.; Bogan, A.E.; Matafonov, D.V.; Froufe, E. Morphological and molecular analyses of Anodontinae species (Bivalvia, Unionidae) of Lake Baikal and Transbaikalia. PLoS ONE 2018, 13, e0194944. [Google Scholar] [CrossRef]
- Hinzmann, M.; Lopes-Lima, M.; Teixeira, A.; Varandas, S.; Sousa, R.; Lopes, A.; Froufe, E.; Machado, J. Reproductive cycle and strategy of Anodonta anatina (L., 1758): Notes on hermaphroditism. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2013, 319, 378–390. [Google Scholar] [CrossRef]
- Reis, J.; Machordom, A.; Araujo, R. Morphological and molecular diversity of Unionidae (Mollusca, Bivalvia) from Portugal. Graellsia 2013, 69, 17–36. [Google Scholar]
- Mezhzherin, S.V.; Yanovich, L.M.; Zhalay, E.I.; Vasilieva, L.A.; Pampura, M.M. Genetic and morphological variability and differentiation of freshwater mussels (Bivavia, Unionidae, Anodontinae) in Ukraine. Vestnik Zoologii 2014, 48, 99–110. [Google Scholar] [CrossRef] [Green Version]
- Araujo, R.; Buckley, D.; Nagel, K.O.; García-Jiménez, R.; Machordom, A. Species boundaries, geographic distribution and evolutionary history of the Western Palaearctic freshwater mussels Unio (Bivalvia: Unionidae). Zool. J. Linn. Soc. 2018, 182, 275–299. [Google Scholar] [CrossRef]
- Araujo, R.; Buckley, D.; Nagel, K.O.; Machordom, A. Potomida littoralis (Bivalvia, Unionidae) evolutionary history: Slow evolution or recent speciation? Zool. J. Linn. Soc. 2017, 179, 277–290. [Google Scholar] [CrossRef] [Green Version]
- Bespalaya, Y.V.; Bolotov, I.N.; Aksenova, O.V.; Gofarov, M.Y.; Kondakov, A.V.; Vikhrev, I.V.; Vinarski, M.V. DNA barcoding reveals invasion of two cryptic Sinanodonta mussel species (Bivalvia: Unionidae) into the largest Siberian River. Limnologica 2018, 69, 94–102. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villesen, P. FaBox: An online toolbox for FASTA sequences. Mol. Ecol. Notes 2007, 7, 965–968. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Froufe, E.; Goncalves, D.V.; Teixeira, A.; Sousa, R.; Varandas, S.; Ghamizi, M.; Zieritz, A.; Lopes-Lima, M. Who lives where? Molecular and morphometric analyses clarify which Unio species (Unionida, Mollusca) inhabit the southwestern Palearctic region. Org. Divers. Evol. 2016, 16, 597–611. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Ho, S.Y.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, 699. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE); IEEE: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Rambaut, A.; Suchard, M.; Drummond, A.J. Tracer v1.6. Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2013. Available online: http://beast.bio.ed.ac.uk/software/tracer/ (accessed on 16 December 2019).
- Cornuet, J.M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.-M.; Estoup, A. DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolotov, I.N.; Aksenova, O.V.; Bespalaya, Y.V.; Gofarov, M.Y.; Kondakov, A.V.; Paltser, I.S.; Stefansson, A.; Travina, O.V.; Vinarski, M.V. Origin of a divergent mtDNA lineage of a freshwater snail species, Radix balthica, in Iceland: Cryptic glacial refugia or a postglacial founder event? Hydrobiologia 2017, 787, 73–98. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver. 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Gomes-dos-Santos, A.; Froufe, E.; Gonçalves, D.V.; Sousa, R.; Prié, V.; Ghamizi, M.; Benaissa, H.; Varandas, S.; Teixeira, A.; Lopes-Lima, M. Freshwater conservation assessments in (semi-)arid regions: Testing river intermittence and buffer strategies using freshwater mussels (Bivalvia, Unionida) in Morocco. Biol. Conserv. 2019, 236, 420–434. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, R.; Sideleva, V.G.; Shedko, S.V.; Goto, A. Broad-scale phylogeography of the Palearctic freshwater fish Cottus poecilopus complex (Pisces: Cottidae). Mol. Phylogenet. Evol. 2008, 48, 1244–1251. [Google Scholar] [CrossRef]
- Sancho, C.; Calle, M.; Peña-Monné, J.L.; Duval, M.; Oliva-Urcia, B.; Pueyo, E.L.; Benito, G.; Moreno, A. Dating the Earliest Pleistocene alluvial terrace of the Alcanadre River (Ebro Basin, NE Spain): Insights into the landscape evolution and involved processes. Quat. Int. 2016, 407, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Feher, Z.; Major, Á.; Krízsik, V. Spatial pattern of intraspecific mitochondrial diversity in the Northern Carpathian endemic spring snail, Bythinella pannonica (Frauenfeld, 1865) (Gastropoda: Hydrobiidae). Org. Divers. Evol. 2013, 13, 569–581. [Google Scholar] [CrossRef]





| Scenarios | Basic Assumptions | Prior Setting of Divergence Time Intervals’ Ranges | Prior Setting of Effective Population Size and Mutation Model |
|---|---|---|---|
| Sc1 | Time scale t3-split of groups IBER, ITAL, AZOV and EUR. | t1 = 105–107 years; t2 = 105–107 years; t3 = 105–107 years; t1 < t2; t2 < t3. | Effective population size: NIBER = 2 × 105–2 × 106 NEUR = 7 × 106–7 × 107 NITAL = 1 × 105–1 × 106 NAZOV = 5 × 105–5 × 106 Evolutionary model: HKY Mutation rate: Uniform COI molecular rate µ= 2.65 × 10−9 substitutions/site/year (rate range: 2.6 × 10−9–2.7 × 10−9 substitutions/site/year) [28] |
| Sc2 | Time scale t3-split of groups AZOV and ITAL, Time scale t2-split of groups IBER and ITAL, Time scale t1-split of groups EUR and IBER. | ||
| Sc3 | Time scale t3-split of groups AZOV and ITAL, Time scale t2-split of groups IBER and AZOV, Time scale t1-split of groups EUR and IBER. |
| IBER | EUR | ITAL | |
|---|---|---|---|
| EUR | 2.56 ± 0.49 | ||
| ITAL | 3.22 ± 0.06 | 3.41 ± 0.07 | |
| AZOV | 2.43 ± 0.05 | 2.35 ± 0.05 | 3.08 ± 0.06 |
| Lineage | N | h | Hd | π | Fu’s FS | Tajima’s D | Mismatch Analysis (Spatial Expansion Model): Estimated τ |
|---|---|---|---|---|---|---|---|
| IBER | 56 | 23 | 0.912 ± 0.022 | 0.858 ± 0.469 | −7.109 * | −0.737 | 6.720 |
| EUR | 168 | 34 | 0.767 ± 0.028 | 0.477 ± 0.280 | −21.078 * | −1.840 * | 0.226 |
| ITAL | 42 | 8 | 0.725 ± 0.058 | 0.477 ± 0.285 | 0.503 | 0.028 | 3.895 |
| AZOV | 58 | 20 | 0.873 ± 0.035 | 0.824 ± 0.452 | −4.375 | −0.320 | 5.769 |
| Direct Approach | Logistic Approach | |||
|---|---|---|---|---|
| Scenario | Posterior Probability, N * = 500 | 95% CI | Posterior Probability, N * = 1 × 105 | 95% CI |
| Sc1 | 0.0740 | 0.0000–0.3035 | 0.0107 | 0.0045–0.0169 |
| Sc2 | 0.4140 | 0.0000–0.8457 | 0.0472 | 0.0358–0.0587 |
| Sc3 | 0.5120 | 0.0739–0.9501 | 0.9421 | 0.9284–0.9557 |
| Parameters | Mean | Median | Qt 5% | Qt 95% |
|---|---|---|---|---|
| Effective population size | ||||
| NIBER | 1.88 × 106 | 1.91 × 106 | 1.67 × 106 | 1.99 × 106 |
| NEUR | 1.03 × 107 | 9.28 × 106 | 7.68 × 106 | 1.56 × 107 |
| NITAL | 8.48 × 105 | 8.80 × 105 | 5.92 × 105 | 9.90 × 105 |
| NAZOV | 3.57 × 106 | 3.64 × 106 | 2.17 × 106 | 4.77 × 106 |
| Divergence time estimation, years | ||||
| ITAL vs. AZOV | 6.11 × 106 | 6.06 × 106 | 3.52 × 106 | 9.01 × 106 |
| AZOV vs. EUR + IBER | 3.61 × 106 | 3.50 × 106 | 1.85 × 106 | 5.73 × 106 |
| EUR vs. IBER | 1.50 × 106 | 1.42 × 106 | 7.69 × 105 | 2.49 × 106 |
| Mutation rate inferred from the mitochondrial COI gene, substitutions/site/year | ||||
| µABC | 2.66 × 10−9 | 2.66 × 10−9 | 2.61 × 10−9 | 2.70 × 10−9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomilova, A.A.; Lyubas, A.A.; Kondakov, A.V.; Vikhrev, I.V.; Gofarov, M.Y.; Kolosova, Y.S.; Vinarski, M.V.; Palatov, D.M.; Bolotov, I.N. Evidence for Plio-Pleistocene Duck Mussel Refugia in the Azov Sea River Basins. Diversity 2020, 12, 118. https://doi.org/10.3390/d12030118
Tomilova AA, Lyubas AA, Kondakov AV, Vikhrev IV, Gofarov MY, Kolosova YS, Vinarski MV, Palatov DM, Bolotov IN. Evidence for Plio-Pleistocene Duck Mussel Refugia in the Azov Sea River Basins. Diversity. 2020; 12(3):118. https://doi.org/10.3390/d12030118
Chicago/Turabian StyleTomilova, Alena A., Artem A. Lyubas, Alexander V. Kondakov, Ilya V. Vikhrev, Mikhail Y. Gofarov, Yulia S. Kolosova, Maxim V. Vinarski, Dmitry M. Palatov, and Ivan N. Bolotov. 2020. "Evidence for Plio-Pleistocene Duck Mussel Refugia in the Azov Sea River Basins" Diversity 12, no. 3: 118. https://doi.org/10.3390/d12030118
APA StyleTomilova, A. A., Lyubas, A. A., Kondakov, A. V., Vikhrev, I. V., Gofarov, M. Y., Kolosova, Y. S., Vinarski, M. V., Palatov, D. M., & Bolotov, I. N. (2020). Evidence for Plio-Pleistocene Duck Mussel Refugia in the Azov Sea River Basins. Diversity, 12(3), 118. https://doi.org/10.3390/d12030118