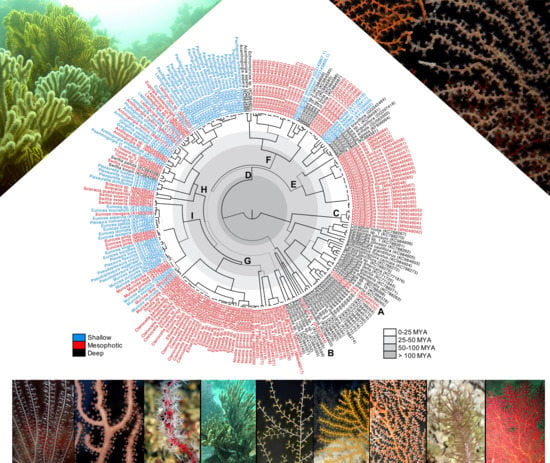
Mesophotic Gorgonian Corals Evolved Multiple Times and Faster Than Deep and Shallow Lineages
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quattrini, A.M.; Georgian, S.E.; Byrnes, L.; Stevens, A.; Falco, R.; Cordes, E.E. Niche divergence by deep-sea octocorals in the genus Callogorgia across the continental slope of the Gulf of Mexico. Mol. Ecol. 2013, 22, 4123–4140. [Google Scholar] [CrossRef]
- Sánchez, J.A.; Dueñas, L.F.; Rowley, S.J.; González, F.L.; Vergara, D.C.; Montaño-Salazar, S.M.; Calixto-Botia, I.; Gómez, C.E.; Abeytia, R.; Colin, P.L.; et al. Gorgonian Corals (39). In Mesophotic Coral Ecosystems; Loya, Y., Puglise, K.A., Bridge, T.C., Eds.; Coral Reefs of the World; Springer Nature AG: Cham, Switzerland, 2019; pp. 727–745. [Google Scholar]
- Velásquez, J.; Sánchez, J.A. Octocoral Species Assembly and Coexistence in Caribbean Coral Reefs. PLoS ONE 2015, 10, e0129609. [Google Scholar]
- Sánchez, J.A. Diversity and evolution of octocoral animal forests at both sides of tropical America. In Marine Animal Forests: The Ecology of Benthic Biodiversity Hotspots; Springer: Cham, Switzerland, 2017; pp. 111–143. [Google Scholar]
- Bates, A.E.; Cooke, R.S.C.; Duncan, M.I.; Edgar, G.J.; Bruno, J.F.; Benedetti-Cecchi, L.; Côté, I.M.; Lefcheck, J.S.; Costello, M.J.; Barrett, N.; et al. Climate resilience in marine protected areas and the ‘Protection Paradox’. Biol. Conserv. 2019, 236, 305–314. [Google Scholar] [CrossRef]
- Stanley, G.D. The evolution of modern corals and their early history. Earth Sci. Rev. 2003, 60, 195–225. [Google Scholar] [CrossRef]
- Prada, C.; Weil, E.; Yoshioka, P.M. Octocoral bleaching during unusual thermal stress. Coral Reefs 2010, 29, 41–45. [Google Scholar] [CrossRef]
- Gómez, C.E.; Paul, V.J.; Ritson-Williams, R.; Muehllehner, N.; Langdon, C.; Sánchez, J.A. Responses of the tropical gorgonian coral Eunicea fusca to ocean acidification conditions. Coral Reefs 2015, 34, 451–460. [Google Scholar] [CrossRef]
- Kocurko, M.J. Shalow-water Octocorallia and related submarine lithification, San Andres island, Colombia. Tex. J. Sci. 1987, 39, 349–365. [Google Scholar]
- Etnoyer, P.; Wirshing, H.; Sánchez, J.A. Rapid assessment of octocoral diversity and habitat on Saba Bank, Netherlands Antilles. PLoS ONE 2010, 5, e10668. [Google Scholar] [CrossRef] [Green Version]
- Pérez, C.D.; de Neves, B.M.; Cordeiro, R.T.; Williams, G.C.; Cairns, S.D. Diversity and Distribution of Octocorallia. In The Cnidaria, Past, Present and Future; Springer: Cham, Switzerland, 2016; pp. 109–123. ISBN 978-3-319-31303-0. [Google Scholar]
- Cordeiro, R.T.; Neves, B.M.; Rosa-Filho, J.S.; Pérez, C.D. Mesophotic coral ecosystems occur offshore and north of the Amazon River. Bull. Mar. Sci. 2015, 91, 491–510. [Google Scholar] [CrossRef]
- Grasshoff, M. The genus Leptogorgia (Octocorallia: Gorgoniidae) in West Africa. Atlantide Rep. 1988, 14, 91–147. [Google Scholar]
- Devictor, S.T.; Morton, S.L. Identification guide to the shallow water (0–200 m) octocorals of the South Atlantic Bight. Zootaxa 2010, 2599, 1–62. [Google Scholar] [CrossRef]
- Perez, C.D.; Neves, B.M.; Oliveira, D.H. New records of octocorals(Cnidaria: Anthozoa) from the Brazilian coast. Aquat. Biol. 2011, 13, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Cerrano, C.; Bastari, A.; Calcinai, B.; Di Camillo, C.; Pica, D.; Puce, S.; Valisano, L.; Torsani, F. Temperate mesophotic ecosystems: Gaps and perspectives of an emerging conservation challenge for the Mediterranean Sea. Eur. Zool. J. 2019, 86, 370–388. [Google Scholar] [CrossRef] [Green Version]
- Gori, A.; Grinyó, J.; Dominguez-Carrió, C.; Ambroso, S.; López-González, P.J.; Gili, J.-M.; Bavestrello, G.; Bo, M. 20 Gorgonian and Black Coral Assemblages in Deep Coastal Bottoms and Continental Shelves of the Mediterranean Sea. In Mediterranean Cold-Water Corals: Past, Present and Future: Understanding the Deep-Sea Realms of Coral; Orejas, C., Jiménez, C., Eds.; Coral Reefs of the World; Springer International Publishing: Cham, Switzerland, 2019; pp. 245–248. ISBN 978-3-319-91608-8. [Google Scholar]
- Kahng, S.E.; Akkaynak, D.; Shlesinger, T.; Hochberg, E.J.; Wiedenmann, J.; Tamir, R.; Tchernov, D. Light, temperature, photosynthesis, heterotrophy, and the lower depth limits of mesophotic coral ecosystems. In Mesophotic Coral Ecosystems; Springer: Cham, Switzerland, 2019; pp. 801–828. [Google Scholar]
- Rocha, L.A.; Pinheiro, H.T.; Shepherd, B.; Papastamatiou, Y.P.; Luiz, O.J.; Pyle, R.L.; Bongaerts, P. Mesophotic coral ecosystems are threatened and ecologically distinct from shallow water reefs. Science 2018, 361, 281–284. [Google Scholar] [CrossRef] [Green Version]
- Quattrini, A.M.; Etnoyer, P.J.; Doughty, C.; English, L.; Falco, R.; Remon, N.; Rittinghouse, M.; Cordes, E.E. A phylogenetic approach to octocoral community structure in the deep Gulf of Mexico. Deep Sea Res. Part II Top. Stud. Oceanogr. 2014, 99, 92–102. [Google Scholar] [CrossRef]
- Soler-Hurtado, M.M.; López-González, P.J.; Machordom, A. Molecular phylogenetic relationships reveal contrasting evolutionary patterns in Gorgoniidae (Octocorallia) in the Eastern Pacific. Mol. Phylogenet. Evol. 2017, 111, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Vargas, S.; Guzman, H.M.; Breedy, O.; Wörheide, G. Molecular phylogeny and DNA barcoding of tropical eastern Pacific shallow-water gorgonian octocorals. Mar. Biol. 2014, 161, 1027–1038. [Google Scholar] [CrossRef]
- Poliseno, A.; Feregrino, C.; Sartoretto, S.; Aurelle, D.; Wörheide, G.; McFadden, C.S.; Vargas, S. Comparative mitogenomics, phylogeny and evolutionary history of Leptogorgia (Gorgoniidae). Mol. Phylogenet. Evol. 2017, 115, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Bilewitch, J.P.; Ekins, M.; Hooper, J.; Degnan, S.M. Molecular and morphological systematics of the Ellisellidae (Coelenterata: Octocorallia): Parallel evolution in a globally distributed family of octocorals. Mol. Phylogenet. Evol. 2014, 73, 106–118. [Google Scholar] [CrossRef]
- Gonzalez-Zapata, F.L.; Bongaerts, P.; Ramírez-Portilla, C.; Adu-Oppong, B.; Walljasper, G.; Reyes, A.; Sánchez, J.A. Holobiont Diversity in a Reef-Building Coral over Its Entire Depth Range in the Mesophotic Zone. Front. Mar. Sci. 2018, 5, 29. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Zapata, F.L.; Gómez-Osorio, S.; Sánchez, J.A. Conspicuous endolithic algal associations in a mesophotic reef-building coral. Coral Reefs 2018, 37, 705–709. [Google Scholar] [CrossRef]
- Sánchez, J.A.; González-Zapata, F.L.; Dueñas, L.F.; Andrade, J.; Pico-Vargas, A.L.; Vergara, D.C.; Sarmiento, A.; Bolaños, N. Corals in the Mesophotic Zone (40–115 m) at the Barrier Reef Complex From San Andrés Island (Southwestern Caribbean). Front. Mar. Sci. 2019, 6, 536. [Google Scholar] [CrossRef] [Green Version]
- Coffroth, M.A.; Lasker, H.R.; Diamond, M.E.; Bruenn, J.A.; Bermingham, E. DNA fingerprints of a gorgonian coral: A method for detecting clonal structure in a vegetative species. Mar. Biol. 1992, 114, 317–325. [Google Scholar] [CrossRef]
- Ardila, N.E.; Giribet, G.; Sánchez, J.A. A time-calibrated molecular phylogeny of the precious corals: Reconciling discrepancies in the taxonomic classification and insights into their evolutionary history. BMC Evol. Biol. 2012, 12, 246. [Google Scholar] [CrossRef] [Green Version]
- McFadden, C.S.; Sánchez, J.A.; France, S.C. Molecular Phylogenetic Insights into the Evolution of Octocorallia: A Review. Integr. Comp. Biol. 2010, 50, 389–410. [Google Scholar] [CrossRef] [Green Version]
- Thorne, J.L.; Kishino, H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002, 51, 689–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzʹmicheva, E.I. Verkhnemelovye I Paleogenovye Korally SSSR; Nauka: Moscow, Russia, 1987. [Google Scholar]
- Hayward, B.W. Lower Miocene corals from the Waitakere Ranges, North Auckland, New Zealand. J. R. Soc. N. Z. 1977, 7, 99–111. [Google Scholar] [CrossRef] [Green Version]
- Kocurko, M.J.; Kocurko, D.J. Fossil Octocorallia of the Red Bluff Formation, lower Oligocene, Mississippi. J. Paleontol. 1992, 66, 594–602. [Google Scholar] [CrossRef]
- FitzJohn, R.G. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 2012, 3, 1084–1092. [Google Scholar] [CrossRef]
- Anderson, D.R. Model Based Inference in the Life Sciences: A Primer on Evidence; Springer Science+ Business Media, LLC: New York, NY, USA, 2008. [Google Scholar]
- Huelsenbeck, J.P.; Nielsen, R.; Bollback, J.P. Stochastic Mapping of Morphological Characters. Syst. Biol. 2003, 52, 131–158. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- McFadden, C.S.; France, S.C.; Sánchez, J.A.; Alderslade, P. A molecular phylogenetic analysis of the Octocorallia (Cnidaria: Anthozoa) based on mitochondrial protein-coding sequences. Mol. Phylogenet. Evol. 2006, 41, 513–527. [Google Scholar] [CrossRef]
- Aguilar, C.; Sánchez, J.A. Phylogenetic hypotheses of gorgoniid octocorals according to ITS2 and their predicted RNA secondary structures. Mol. Phylogenet. Evol. 2007, 43, 774–786. [Google Scholar] [CrossRef]
- Wirshing, H.H.; Messing, C.G.; Douady, C.J.; Reed, J.; Stanhope, M.J.; Shivji, M.S. Molecular evidence for multiple lineages in the gorgonian family Plexauridae (Anthozoa: Octocorallia). Mar. Biol. 2005, 147, 497–508. [Google Scholar] [CrossRef]
- Holm, J.B.; Heidelberg, K.B. Microbiomes of Muricea californica and M. fruticosa: Comparative analyses of two co-occurring Eastern Pacific octocorals. Front. Microbiol. 2016, 7, 917. [Google Scholar] [CrossRef]
- Breedy, O.; Guzman, H.M. A New Species of the Genus Eugorgia (Cnidaria: Octocorallia: Gorgoniidae) from Mesophotic Reefs in the Eastern Pacific. Bull. Mar. Sci. 2013, 89, 735–743. [Google Scholar] [CrossRef]
- Cordeiro, R.T.S.; McFadden, C.S.; Sanchez, J.A.; Pérez, C.D. Revision of the genus Plexaurella Kölliker, 1865 (Anthozoa: Octocorallia) and resurrection of Plexaurellidae Verrill, 1912 new rank. Invertebr. Syst. 2021, 35, 892–921. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Baums, I.B.; Shank, T.M.; Morrison, C.L.; Cordes, E.E. Testing the depth-differentiation hypothesis in a deepwater octocoral. Proc. R. Soc. B Biol. Sci. 2015, 282, 20150008. [Google Scholar] [CrossRef] [Green Version]
- Bayer, F.M. The shallow-water Octocorallia of the West Indian region. Stud. Fauna Curacao Other Caribb. Isl. 1961, 12, 1–373. [Google Scholar]
- Quattrini, A.M.; Rodríguez, E.; Faircloth, B.C.; Cowman, P.F.; Brugler, M.R.; Farfan, G.A.; Hellberg, M.E.; Kitahara, M.V.; Morrison, C.L.; Paz-García, D.A. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 2020, 4, 1531–1538. [Google Scholar] [CrossRef]
- McFadden, C.S.; Quattrini, A.M.; Brugler, M.R.; Cowman, P.F.; Dueñas, L.F.; Kitahara, M.V.; Paz-García, D.A.; Reimer, J.D.; Rodríguez, E. Phylogenomics, Origin, and Diversification of Anthozoans (Phylum Cnidaria). Syst. Biol. 2021, 70, 635–647. [Google Scholar] [CrossRef]
- Bayer, F.M. Thelogorgia, a new genus of gorgonacean octocorals, with descriptions of four new species from the western Atlantic. Bull. Mar. Sci. 1991, 49, 506–537. [Google Scholar]
- Sánchez, J.A. Black coral-octocoral distribution patterns in Imelda bank, a deep-water reef, Colombia, Caribbean sea. Bull. Mar. Sci. 1999, 65, 215–225. [Google Scholar]
- Quattrini, A.M.; Faircloth, B.C.; Dueñas, L.F.; Bridge, T.C.L.; Brugler, M.R.; Calixto-Botía, I.F.; DeLeo, D.M.; Forêt, S.; Herrera, S.; Lee, S.M.Y.; et al. Universal target-enrichment baits for anthozoan (Cnidaria) phylogenomics: New approaches to long-standing problems. Mol. Ecol. Resour. 2018, 18, 281–295. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Gómez, C.E.; Cordes, E.E. Environmental filtering and neutral processes shape octocoral community assembly in the deep sea. Oecologia 2017, 183, 221–236. [Google Scholar] [CrossRef]
- Bongaerts, P.; Frade, P.R.; Hay, K.B.; Englebert, N.; Latijnhouwers, K.R.; Bak, R.P.; Vermeij, M.J.; Hoegh-Guldberg, O. Deep down on a Caribbean reef: Lower mesophotic depths harbor a specialized coral-endosymbiont community. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- Kahng, S.E.; Copus, J.M.; Wagner, D. Recent advances in the ecology of mesophotic coral ecosystems (MCEs). Curr. Opin. Environ. Sustain. 2014, 7, 72–81. [Google Scholar] [CrossRef]
- Stolarski, J.; Kitahara, M.V.; Miller, D.J.; Cairns, S.D.; Mazur, M.; Meibom, A. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol. Biol. 2011, 11, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldwin, C.C.; Tornabene, L.; Robertson, D.R. Below the Mesophotic. Sci. Rep. 2018, 8, 4920. [Google Scholar] [CrossRef] [PubMed]
- Calixto-Botía, I.; Sánchez, J.A. A case of modular phenotypic plasticity in the depth gradient for the gorgonian coral Antillogorgia bipinnata (Cnidaria: Octocorallia). BMC Evol. Biol. 2017, 17, 55. [Google Scholar] [CrossRef] [Green Version]
- Prada, C.; Hellberg, M.E. Long prereproductive selection and divergence by depth in a Caribbean candelabrum coral. Proc. Natl. Acad. Sci. USA 2013, 110, 3961–3966. [Google Scholar] [CrossRef] [Green Version]
- Ament-Velásquez, S.L.; Breedy, O.; Cortés, J.; Guzman, H.M.; Wörheide, G.; Vargas, S. Homoplasious colony morphology and mito-nuclear phylogenetic discordance among Eastern Pacific octocorals. Mol. Phylogenet. Evol. 2016, 98, 373–381. [Google Scholar] [CrossRef]
- Grajales, A.; Aguilar, C.; Sánchez, J.A. Phylogenetic reconstruction using secondary structures of Internal Transcribed Spacer 2 (ITS2, rDNA): Finding the molecular and morphological gap in Caribbean gorgonian corals. BMC Evol. Biol. 2007, 7, 90. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, J.A.; MacFadden, C.S.; France, S.C.; Lasker, H.R. Molecular phylogenetic analyses of shallow-water Caribbean octocorals. Mar. Biol. 2003, 142, 975–987. [Google Scholar] [CrossRef]
- Wirshing, H.H.; Baker, A.C. Molecular and Morphological Species Boundaries in the Gorgonian Octocoral Genus Pterogorgia (Octocorallia: Gorgoniidae). PLoS ONE 2015, 10, e0133517. [Google Scholar] [CrossRef]
- Prada, C.; Schizas, N.V.; Yoshioka, P.M. Phenotypic plasticity or speciation? A case from a clonal marine organism. BMC Evol. Biol. 2008, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Prada, C.; Hellberg, M.E. Speciation-by-depth on coral reefs: Sympatric divergence with gene flow or cryptic transient isolation? J. Evol. Biol. 2021, 34, 128–137. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez, J.A.; González-Zapata, F.L.; Prada, C.; Dueñas, L.F. Mesophotic Gorgonian Corals Evolved Multiple Times and Faster Than Deep and Shallow Lineages. Diversity 2021, 13, 650. https://doi.org/10.3390/d13120650
Sánchez JA, González-Zapata FL, Prada C, Dueñas LF. Mesophotic Gorgonian Corals Evolved Multiple Times and Faster Than Deep and Shallow Lineages. Diversity. 2021; 13(12):650. https://doi.org/10.3390/d13120650
Chicago/Turabian StyleSánchez, Juan A., Fanny L. González-Zapata, Carlos Prada, and Luisa F. Dueñas. 2021. "Mesophotic Gorgonian Corals Evolved Multiple Times and Faster Than Deep and Shallow Lineages" Diversity 13, no. 12: 650. https://doi.org/10.3390/d13120650
APA StyleSánchez, J. A., González-Zapata, F. L., Prada, C., & Dueñas, L. F. (2021). Mesophotic Gorgonian Corals Evolved Multiple Times and Faster Than Deep and Shallow Lineages. Diversity, 13(12), 650. https://doi.org/10.3390/d13120650