Managing Genetic Diversity and Representation in Banksia marginata (Proteaceae) Seed Production Areas Used for Conservation and Restoration
Abstract
:1. Introduction
2. Materials and Methods
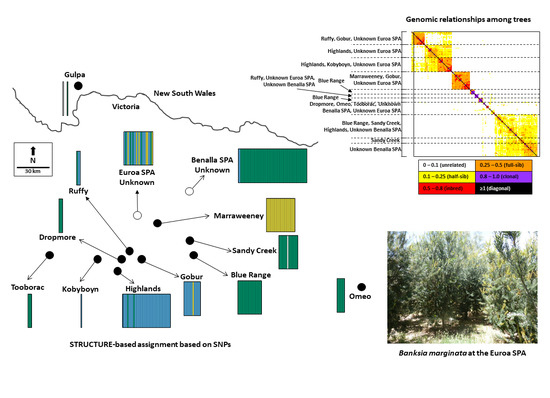
2.1. Species Background, Study Sites and Sampling
2.2. DNA Extraction and Genotyping
2.3. Genetic Diversity and Relatedness in Remnant and SPA Populations
2.4. Population Genetic Structure
3. Results
3.1. Genetic Diversity and Relatedness in Wild and SPA Populations
3.2. Population Genetic Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPBES. Summary for Policymakers of the Thematic Assessment Report on Land Degradation and Restoration of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; IPBES Secretariat: Bonn, Germany, 2018. [Google Scholar]
- Mortlock, W. Local seed for revegetation: Where will all that seed come from? Ecol. Manag. Restor. 2000, 1, 93–101. [Google Scholar] [CrossRef]
- Merritt, D.J.; Dixon, K.W. Restoration Seed Banks—A Matter of Scale. Science 2011, 332, 424–425. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, L.; Lowe, A.; Coates, D.; Cunningham, S.; McDonald, M.; Vesk, P.; Yates, C. Seed supply for broadscale restoration: Maximizing evolutionary potential. Evol. Appl. 2008, 1, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, L.M.; Jones, T.A.; Smith, F.; North, T.; Guja, L. Maximizing seed resources for restoration in an uncertain future. Bioscience 2016, 66, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Broadhurst, L.; Hopley, T.; Li, L.; Begley, J. A genetic assessment of seed production areas (SPAs) for restoration. Conserv. Genet. 2017, 18, 1257–1266. [Google Scholar] [CrossRef]
- Gordon, D.R.; Rice, K.J. Patterns of differentiation in wiregrass (Aristida beyrichiana): Implications for restoration. Restor. Ecol. 1998, 6, 166–174. [Google Scholar] [CrossRef]
- Avise, J.C. A role for molecular genetics in the recognition and conservation of endangered species. Trends Ecol. Evol. 1989, 4, 279–281. [Google Scholar] [CrossRef] [Green Version]
- Helenurm, K. Outplanting and differential source population success in Lupinus guadalupensis. Conserv. Biol. 1998, 12, 118–127. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetics consequences of small population size: Implications for plant conservation. Ann. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Young, A.G.; Boyle, T.J.B.; Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996, 11, 413–417. [Google Scholar] [CrossRef]
- Hobbs, R.J.; Higgs, E.; Harris, J.A. Novel ecosystems: Implications for conservation and restoration. Trends Ecol. Evol. 2009, 24, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.A. Ecologically appropriate plant materials for functional restoration of rangelands. J. Sust. Forest. 2014, 33, S93–S103. [Google Scholar] [CrossRef]
- Jones, T.A. Ecologically appropriate plant materials for restoration applications. BioScience 2013, 63, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Vander Mijnsbrugge, K.; Bischoff, A.; Smith, B. A question of origin: Where and how to collect seed for ecological restoration. Basic Appl. Ecol. 2010, 11, 300–311. [Google Scholar] [CrossRef] [Green Version]
- Butcher, P.; Williams, E. Variation in outcrossing rates and growth in Eucalyptus camaldulensis from the Petford region, Queensland; evidence of outbreeding depression. Silvae Genet. 2002, 51, 6–12. [Google Scholar]
- Jones, R.; Vaillancourt, R.; Gore, P.; Potts, B. Genetic control of flowering time in Eucalyptus globulus ssp. globulus. Tree Genet. Genomes 2011, 7, 1209–1218. [Google Scholar] [CrossRef] [Green Version]
- Aguilar, R.; Ashworth, L.; Galetto, L.; Aizen, M.A. Plant reproduction susceptibility to habitat fragmentation: Review and synthesis through a meta-analysis. Ecol. Lett. 2006, 9, 968–980. [Google Scholar] [CrossRef]
- Broadhurst, L.; Fifield, G.; Vanzella, B.; Pickup, M. Evaluating the evolutionary potential of two Yellow Box (Eucalyptus melliodora A.Cunn. ex Schauer, Myrtaceae) seed production areas. Aust. J. Bot. 2015, 63, 455–466. [Google Scholar] [CrossRef]
- Dolan, R.W.; Marr, D.L.; Schnabel, A. Capturing genetic variation during ecological restorations: An example from Kankakee Sands in Indiana. Restor. Ecol. 2008, 16, 386–396. [Google Scholar] [CrossRef]
- Fant, J.B.; Holmstrom, R.M.; Sirkin, E.; Etterson, J.R.; Masi, S. Genetic structure of threatened native populations and propagules used for restoration in a clonal species, American Beachgrass (Ammophila breviligulata Fern.). Restor. Ecol. 2008, 16, 594–603. [Google Scholar] [CrossRef]
- Dyer, A.R.; Knapp, E.E.; Rice, K.J. Unintentional selection and genetic changes in native perennial grass populations during commercial seed production. Ecol. Restor. 2016, 34, 39–48. [Google Scholar] [CrossRef]
- Kettle, C.J.; Ennos, R.A.; Jaffre, T.; Gardner, M.; Hollingsworth, P.M. Cryptic genetic bottlenecks during restoration of an endangered tropical conifer. Biol. Conserv. 2008, 141, 1953–1961. [Google Scholar] [CrossRef]
- Broadhurst, L.; Breed, M.; Lowe, A.; Bragg, J.; Catullo, R.; Coates, D.; Encinas-Viso, F.; Gellie, N.; James, E.; Krauss, S.; et al. Genetic diversity and structure of the Australian flora. Divers. Distrib. 2017, 23, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996, 351, 1291–1298. [Google Scholar] [CrossRef]
- Nybom, H.; Bartish, I.V. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect. Plant Ecol. Evol. Syst. 2000, 3, 93–114. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Hamrick, J.L. Response of forest trees to global environmental changes. Forest Ecol. Manag. 2004, 197, 323–335. [Google Scholar] [CrossRef]
- Hancock, N.; Gibson Roy, P.; Driver, M.; Broadhurst, L. The Australian Native Seed Survey Report; Australian Network for Plant Conservation: Canberra, Australia, 2020. [Google Scholar]
- Broadhurst, L.; Driver, M.; Guja, L.; North, T.; Vanzella, B.; Fifield, G.; Bruce, S.; Taylor, D.; Bush, D. Seeding the Future – the issues of supply and demand in restoration. Ecol. Manag. Restor. 2015, 16, 29–32. [Google Scholar] [CrossRef]
- Taylor, A.; Hopper, S.D. Banksia Atlas; Australian Government Publishing Service: Canberra, Australia, 1991.
- Collins, K.; George, A. Banksias; Bloomings Books: Melbourne, Australia, 2008. [Google Scholar]
- George, A.S. The Banksia Book; Kangaroo Press: Sydney, Australia, 1984. [Google Scholar]
- George, A.S. The genus Banksia L.f. (Proteaceae). Nuytsia 1981, 3, 239–473. [Google Scholar]
- Vaughton, G.; Ramsey, M. Selfed seed set and inbreeding depression in obligate seeding populations of Banksia marginata. Proceed. Linnean Soc. NSW 2006, 127, 19–25. [Google Scholar]
- Charlesworth, D.; Charlesworth, B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987, 18, 237–268. [Google Scholar] [CrossRef]
- Vaughton, G.; Ramsey, M. Sources and consequences of seed mass variation in Banksia marginata (Proteaceae). J. Ecol. 1998, 86, 563–573. [Google Scholar] [CrossRef]
- Specht, R.; Rayson, P.; Jackman, M. Dark Island heath (Ninety-mile Plain, South Australia). VI. Pyric succession: Changes in composition, coverage, dry weight, and mineral nutrient status. Aust. J. Bot. 1958, 6, 59–88. [Google Scholar] [CrossRef]
- Ford, H.A.; Paton, D.C.; Forde, N. Birds as pollinators of Australian plants. N. Z. J. Bot. 1979, 17, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Lepschi, B. Food of some birds in southern Australia: Additions to Barker & Vestjens, Part 2. EMU 1997, 97, 84–87. [Google Scholar]
- Duncan, A.; Taylor, R. Occurrence of pygmy possums, Cercartetus lepidus and C. nanus, and their nest sites in logged and unlogged dry and wet eucalypt forest in Tasmania. Aust. For. 2001, 64, 159–164. [Google Scholar] [CrossRef]
- Hord, H.A.; Paton, D.C. Partitioning of nectar sources in an Australian honeyeater community. Aust. J. Ecol. 1982, 7, 149–159. [Google Scholar] [CrossRef]
- Swart, H.J. Australian leaf-inhabiting fungi XXIX. Some ascomycetes on Banksia. Trans. Brit. Mycol. Soc. 1988, 91, 453–465. [Google Scholar] [CrossRef]
- Blundell, M.J.; Phongkham, T.; Gale, K.R. A novel enzyme-linked immunosorbent assay for the detection of WX-B1 null wheat lines using a monclonal antibody and its application for the detection of marker heterogeneity within commercial cultivars. J. Cereal Sci. 2010, 52, 187–191. [Google Scholar] [CrossRef]
- Usher, A.V.; Ayre, D.J.; Whelan, R.J. Microsatellites for eastern Australian Banksia species. Mol. Ecol. Notes 2005, 5, 821–823. [Google Scholar] [CrossRef]
- He, T.; Krauss, S.L.; Lamont, B.B. Polymorphic microsatellite DNA markers for Banksia hookeriana (Proteaceae). Mol. Ecol. Resour. 2008, 8, 1515–1517. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, M.; Houliston, G.J.; Haddadchi, A.; Gross, C.L. Cost-effective microsatellite markers for Banksia integrifolia (Proteaceae). Appl. Plant Sci. 2013, 1, 1200130. [Google Scholar] [CrossRef] [PubMed]
- Nistelberger, H.M.; Llorens, T.M.; Byrne, M. Isolation and characterization of microsatellites in the woody shrub, Banksia sphaerocarpa var. caesia (Proteaceae). Mol. Ecol. Resour. 2009, 9, 148–149. [Google Scholar] [CrossRef] [PubMed]
- Merwin, L.; He, T.; Krauss, S.L. Isolation and characterization of polymorphic microsatellite DNA markers for Banksia candolleana (Proteaceae). Conserv. Genet. Resour. 2010, 2, 345–347. [Google Scholar] [CrossRef]
- He, T.; Krauss, S.; Lamont, B.B. Polymorphic microsatellite DNA markers for Banksia attenuata (Proteaceae). Mol. Ecol. Notes 2007, 7, 1329–1331. [Google Scholar] [CrossRef]
- Broadhurst, L.M. Genetic diversity and population genetic structure in fragmented Allocasuarina verticillata (Allocasuarinaceae) - implications for restoration. Aust. J. Bot. 2011, 59, 770–780. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, 54. [Google Scholar] [CrossRef] [Green Version]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics; Springer: Berlin/Heidelberg, Germany, 2012; pp. 67–89. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 228–295. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3). Available online: http://www2.unil.ch/izea/softwares/fstat.html (accessed on 5 February 2002).
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. dartr: An R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [PubMed]
- Queller, D.C.; Goodnight, K.F. Estimating relatedness using genetic markers. Evolution 1989, 43, 258–275. [Google Scholar] [CrossRef]
- Wang, J. Coancestry: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 2010, 11, 141–145. [Google Scholar] [CrossRef]
- Wang, J. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 2007, 89, 135–153. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.; vonHoldt, B. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef] [PubMed]
- Nickolas, H.; Harrison, P.A.; Tilyard, P.; Vaillancourt, R.E.; Potts, B.M. Inbreeding depression and differential maladaptation shape the fitness trajectory of two co-occurring Eucalyptus species. Ann. For. Sci. 2019, 76, 10. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.D.; Nitschke, C.; Weeks, A.R.; Weatherly, W.L.; Heyes, S.D.; Sinclair, S.J.; Holland, O.J.; Stevenson, A.; Broadhurst, L.; Hoebee, S.E.; et al. Genetic data and climate niche suitability models highlight the vulnerability of a functionally important plant species from south-eastern Australia. Evol. Appl. 2020, 13, 2014–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyes, S.D.; Sinclair, S.J.; Hoebee, S.E.; Morgan, J.W. How widespread are recruitment bottlenecks in fragmented populations of the savanna tree Banksia marginata (Proteaceae)? Plant Ecol. 2020, 221, 545–557. [Google Scholar] [CrossRef]
- Millar, M.A.; Byrne, M. Variable clonality and genetic structure among disjunct populations of Banksia mimica. Conserv. Genet. 2020, 21, 803–818. [Google Scholar] [CrossRef]
- Nistelberger, H.M.; Byrne, M.; Coates, D.; Roberts, J.D. Phylogeography and population differentiation in terrestrial island populations of Banksia arborea (Proteaceae). Biol. J. Linn. Soc. Lond. 2015, 114, 860–872. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, A.L.; Dyer, R.J.; Nevill, P.G.; Sinclair, E.A.; Krauss, S.L. Wide outcrossing provides functional connectivity for new and old Banksia populations within a fragmented landscape. Oecologia 2019, 190, 255–268. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, D.; Binks, R.M.; Coates, D.J.; Byrne, M. Conservation genomics of range disjunction in a global biodiversity hotspot: A case study of Banksia biterax (Proteaceae) in southwestern Australia. Biol. J. Linn. Soc. Lond. 2019, 127, 390–406. [Google Scholar] [CrossRef]
- Robins, T.P.; Binks, R.M.; Byrne, M.; Hopper, S.D. Contrasting patterns of population divergence on young and old landscapes in Banksia seminuda (Proteaceae), with evidence for recognition of subspecies. Biol. J. Linn. Soc. Lond. 2020. [Google Scholar] [CrossRef]
- Vranckx, G.; Jacquemyn, H.; Muys, B.; Honnay, O. Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv. Biol. 2012, 26, 228–237. [Google Scholar] [CrossRef] [PubMed]
- González, A.V.; Gómez-Silva, V.; Ramírez, M.J.; Fontúrbel, F.E. Meta-analysis of the differential effects of habitat fragmentation and degradation on plant genetic diversity. Conserv. Biol. 2020, 34, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, P.; Hensen, I.; Renison, D.; Wesche, K.; Ploch, S.; Rondan Duenas, J.; Cocucci, A.; Jung, K. Biparental inbreeding depression, genetic relatedness and progeny vigour in a wind-pollinated treeline species in Argentina. Plant Ecol. 2009, 205, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Seltmann, P.; Renison, D.; Cocucci, A.; Hensen, I.; Jung, K. Fragment size, pollination efficiency and reproductive success in natural populations of wind-pollinated Polylepis australis (Rosaceae) trees. Flora 2007, 202, 547–554. [Google Scholar] [CrossRef]
- Jump, A.S.; Penuelas, J. Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proc. Natl. Acad. Sci. USA 2006, 103, 8096–8100. [Google Scholar] [CrossRef] [Green Version]
- Ford, H.A.; Barrett, G.W.; Saunders, D.A.; Recher, H.F. Why have birds in the woodlands of Southern Australia declined? Biol. Conserv. 2001, 97, 71–88. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Lane, P.; Westgate, M.; Scheele, B.C.; Foster, C.; Sato, C.; Ikin, K.; Crane, M.; Michael, D.; Florance, D.; et al. Tests of predictions associated with temporal changes in Australian bird populations. Biol. Conserv. 2018, 222, 212–221. [Google Scholar] [CrossRef]
- Young, A.G.; Pickup, M. Low S-allele numbers limit mate availability, reduce seed set and skew fitness in small populations of a self-incompatible plant. J. Appl. Ecol. 2010, 47, 541–548. [Google Scholar] [CrossRef]
- Broadhurst, L.M.; Young, A.G. Reproductive constraints to the long-term persistence of fragmented Acacia dealbata (Mimosaceae) populations in southeast Australia. Biol. Conserv. 2006, 133, 512–526. [Google Scholar] [CrossRef]
- Carthew, S.M.; Whelan, R.J.; Ayre, D.J. Experimental Confirmation of Preferential Outcrossing in Banksia. Int. J. Plant Sci. 1996, 157, 615–620. [Google Scholar] [CrossRef]
- Pryor, L.D. Biology of Eucalypts; Edward Arnold: London, UK, 1976. [Google Scholar]
- Heliyanto, B.; Veneklaas, E.J.; Lambers, H.; Krauss, S.L. Preferential outcrossing in Banksia ilicifolia (Proteaceae). Aust. J. Bot. 2005, 53, 164–170. [Google Scholar] [CrossRef]
- Moran, G.; Bell, C.; Griffin, A.R. Reduction in levels of inbreeding in a seed orchard of Eucalyptus regnans F.Muell. compared with natural populations. Silvae Genet. 1989, 38, 32–36. [Google Scholar]
- Ellis, M.F.; Sedgley, M. Floral morphology and breeding system of three species of Eucalyptus, section Bisectaria (Myrtaceae). Aust. J. Bot. 1992, 40, 249–262. [Google Scholar] [CrossRef]
- Griffin, A.R.; Potts, B.M.; Vaillancourt, R.E.; Bell, J.C. Life cycle expression of inbreeding depression in Eucalyptus regnans and inter-generational stability of its mixed mating system. Ann. Bot. 2019, 124, 179–187. [Google Scholar] [CrossRef]
- Wooller, R.; Russell, E.; Renfree, M.; Towers, P. A comparison of seasonal changes in the pollen loads of nectarivorous marsupials [Tarsipes] and birds [Honeyeaters]. Wildl. Res. 1983, 10, 311–317. [Google Scholar] [CrossRef]
- Blake, S.T. Flowering and seeding habits in some species of Banksia. QLD Natur. 1971, 20, 21–24. [Google Scholar]
- Frick, K.M.; Ritchie, A.L.; Krauss, S.L. Field of dreams: Restitution of pollinator services in restored bird-pollinated plant populations. Restor. Ecol. 2014, 22, 832–840. [Google Scholar] [CrossRef]
- Llorens, T.M.; Byrne, M.; Yates, C.J.; Nistelberger, H.M.; Coates, D.J. Evaluating the influence of different aspects of habitat fragmentation on mating patterns and pollen dispersal in the bird-pollinated Banksia sphaerocarpa var. caesia. Mol. Ecol. 2012, 21, 314–328. [Google Scholar] [CrossRef]
- Krauss, S.; He, T.; Barrett, L.; Lamont, B.; Enright, N.; Miller, B.; Hanley, M. Contrasting impacts of pollen and seed dispersal on spatial genetic structure in the bird-pollinated Banksia hookeriana. Heredity 2009, 102, 274–285. [Google Scholar] [CrossRef] [Green Version]
- He, T.; Krauss, S.L.; Lamont, B.B.; Miller, B.P.; Enright, N.J. Long-distance seed dispersal in a metapopulation of Banksia hookeriana inferred from a population allocation analysis of amplified fragment length polymorphism data. Mol. Ecol. 2004, 13, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R. Statistical Methods for Research Workers; Oliver and Boyd: Edinburgh, UK, 1954. [Google Scholar]






| Type | Name | N | Na (s.e.) | Rs (s.e.) * | Ho (s.e.) | UHe (s.e.) | FIS (s.e.) |
|---|---|---|---|---|---|---|---|
| Remnant | Blue Range | 17 | 4.34 (0.99) | 2.16 (0.28) | 0.46 (0.10) | 0.53 (0.09) | 0.03 (0.13) |
| Dropmore | 3 | 2.00 (0.27) | 1.62 (0.19) | 0.38 (0.13) | 0.38 (0.10) | −0.15 (0.21) | |
| Gobur | 11 | 2.88 (0.40) | 1.94 (0.17) | 0.49 (0.12) | 0.47 (0.08) | −0.12 (0.19) | |
| Gulpa† | NA | NA | NA | NA | NA | NA | |
| Highlands | 34 | 4.38 (0.38) | 2.01 (0.17) | 0.55 (0.11) | 0.48 (0.08) | −0.11 (0.12) | |
| Kobyboyn | 1 | 1.63 (0.18) | NA | 0.63 (0.18) | 0.63 (0.18) | NA | |
| Marraweeney | 21 | 3.63 (0.42) | 2.11 (0.16) | 0.66 (0.10) | 0.54 (0.07) | −0.20 (0.14) | |
| Omeo | 5 | 3.50 (0.50) | 2.33 (0.24) | 0.69 (0.12) | 0.59 (0.09) | −0.29 (0.16) | |
| Tooborac | 3 | 2.63 (0.32) | 2.28 (0.23) | 0.38 (0.10) | 0.60 (0.09) | 0.27 (0.15) | |
| Ruffy | 3 | 2.38 (0.26) | 2.07 (0.23) | 0.67 (0.08) | 0.58 (0.05) | −0.47 (0.14) | |
| Sandy Creek | 15 | 3.88 (0.48) | 2.24 (0.15) | 0.63 (0.08) | 0.57 (0.05) | −0.20 (0.10) | |
| Unknown | 72 | 6.75 (0.99) | 2.44 (0.19) | 0.58 (0.08) | 0.64 (0.07) | 0.05 (0.12) | |
| Pooled | Wild | 29 | 6.00 (1.07) | 5.97 (1.07) | 0.55 (0.09) | 0.59 (0.08) | 0.05 (0.12) |
| Euroa SPA | 108 | 6.88 (0.92) | 5.22 (0.71) | 0.56 (0.08) | 0.58 (0.08) | 0.00 (0.11) | |
| Benalla SPA | 50 | 5.88 (0.95) | 5.29 (0.88) | 0.60 (0.08) | 0.62 (0.06) | 0.00 (0.12) |
| Type | Name | N | Rs (s.e.) * | Ho (s.e.) | He (s.e.) | FIS (s.e.) |
|---|---|---|---|---|---|---|
| Remnant | Blue Range | 19 | 1.26 (<0.01) | 0.21 (<0.01) | 0.25 (<0.01) | 0.16 (<0.01) |
| Dropmore | 3 | 1.18 (<0.01) | 0.20 (<0.01) | 0.15 (<0.01) | −0.33 (<0.01) | |
| Gobur | 13 | 1.23 (<0.01) | 0.22 (<0.01) | 0.22 (<0.01) | 0.00 (<0.01) | |
| Gulpa† | 1 | 1.12 (<0.01) | 0.12 (<0.01) | 0.06 (<0.01) | NA | |
| Highlands | 39 | 1.26 (<0.01) | 0.23 (<0.01) | 0.26 (<0.01) | 0.12 (<0.01) | |
| Kobyboyn | 1 | 1.25 (0.01) | 0.25 (<0.01) | 0.13 (<0.01) | NA | |
| Marraweeney | 23 | 1.22 (<0.01) | 0.23 (<0.01) | 0.21 (<0.01) | −0.10 (<0.01) | |
| Omeo | 5 | 1.23 (<0.01) | 0.20 (<0.01) | 0.21 (<0.01) | 0.05 (<0.01) | |
| Tooborac | 4 | 1.26 (<0.01) | 0.21 (<0.01) | 0.22 (<0.01) | 0.05 (<0.01) | |
| Ruffy | 3 | 1.24 (0.01) | 0.24 (<0.01) | 0.20 (<0.01) | −0.20 (<0.01) | |
| Sandy Creek | 15 | 1.25 (<0.01) | 0.22 (<0.01) | 0.24 (<0.01) | 0.17 (<0.01) | |
| Unknown | 83 | 1.31 (<0.01) | 0.23 (<0.01) | 0.30 (<0.01) | 0.23 (<0.01) | |
| Pooled | Wild | 32 | 1.87 (0.005) | 0.22 (<0.01) | 0.29 (<0.01) | 0.24 (<0.01) |
| Euroa SPA | 120 | 1.92 (0.003) | 0.23 (<0.01) | 0.31 (<0.01) | 0.27 (<0.01) | |
| Benalla SPA | 57 | 1.82 (0.006) | 0.23 (<0.01) | 0.28 (<0.01) | 0.17 (<0.01) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broadhurst, L.; Bush, D.; Begley, J. Managing Genetic Diversity and Representation in Banksia marginata (Proteaceae) Seed Production Areas Used for Conservation and Restoration. Diversity 2021, 13, 39. https://doi.org/10.3390/d13020039
Broadhurst L, Bush D, Begley J. Managing Genetic Diversity and Representation in Banksia marginata (Proteaceae) Seed Production Areas Used for Conservation and Restoration. Diversity. 2021; 13(2):39. https://doi.org/10.3390/d13020039
Chicago/Turabian StyleBroadhurst, Linda, David Bush, and Jim Begley. 2021. "Managing Genetic Diversity and Representation in Banksia marginata (Proteaceae) Seed Production Areas Used for Conservation and Restoration" Diversity 13, no. 2: 39. https://doi.org/10.3390/d13020039
APA StyleBroadhurst, L., Bush, D., & Begley, J. (2021). Managing Genetic Diversity and Representation in Banksia marginata (Proteaceae) Seed Production Areas Used for Conservation and Restoration. Diversity, 13(2), 39. https://doi.org/10.3390/d13020039