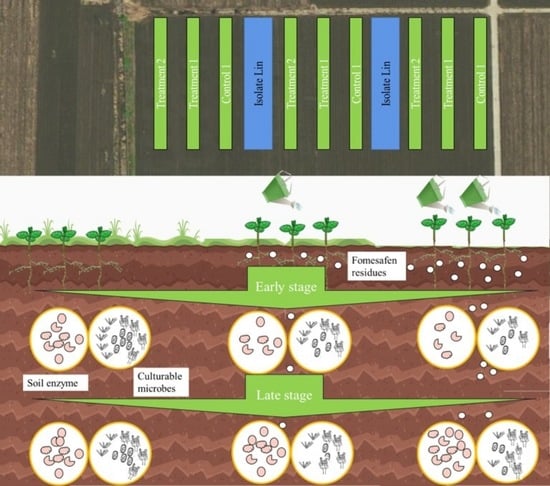
Microbial Community’s Dynamic Response to Fomesafen Usage in Chernozems of Northeast China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil
2.2. Soil Fomesafen Residue Assays
2.3. Soil Enzyme Assays
2.4. Soil Culturable Microbial Analysis
2.5. Soil PLFA Analysis
2.6. Statistical Analysis
3. Results
3.1. Soil Characteristics and Fomesafen Residues
3.2. Soil Enzyme Activities: Catalase, Urease, Saccharase, Acid Phosphatase
3.3. Soil Culturable Microbes’ Biomass: Bacteria, Fungi and Actinobacteria
3.4. Soil PLFA Analysis and Microbial Community Structure Reflected by PLFA Patterns
3.5. Correlation between Soil Microbial Community Composition and Environmental Factors
4. Discussion
4.1. Soil Fomesafen Residue Dynamics
4.2. Effect of Fomesafen on Soil Enzyme Activities
4.3. Effect of Fomesafen on the Biomass of Culturable Bacteria, Fungi, and Actinomycetes in Soil
4.4. Soil PLFA Analysis and Microbial Community Structure Reflected by PLFA Patterns
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Everman, W.J.; Clewis, S.B.; York, A.C.; Wilcut, J.W. Weed control and yield with flumioxazin, fomesafen, and s-metolachlor systems for glufosinate-resistant cotton residual weed management. Weed Technol. 2017, 23, 391–397. [Google Scholar] [CrossRef]
- Costa, A.I.G.; Queiroz, M.E.L.R.; Neves, A.; De Assis, R.C.; Dos Soares, C.E.S.; Da Silva, A.A.; D’Antonino, L.; De Oliveira, A.F.; Bellato, C.R. Mobility and persistence of the herbicide fomesafen in soils cultivated with bean plants using SLE/LTP and HPLC/DAD. Environ. Sci. Pollut. Res. 2015, 22, 3457–3466. [Google Scholar] [CrossRef]
- Williams, M.M.; Nelson, R.L. Vegetable soybean tolerance to bentazon, fomesafen, imazamox, linuron, and sulfentrazone. Weed Technol. 2014, 28, 601–607. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, M.; An, S.; Xiong, B.; Li, H.; Cui, C.Z.; Lin, K.F. Ecotoxicological effects of decabromodiphenyl ether and cadmium contamination on soil microbes and enzymes. Ecotoxicol. Environ. Saf. 2012, 82, 71–79. [Google Scholar] [CrossRef]
- Guo, H.; Yao, J.; Cai, M.; Qian, Y.; Yue, G.; Richnow, H.H.; Blake, R.E.; Doni, S.; Ceccanti, B. Effects of petroleum contamination on soil microbial numbers, metabolic activity and urease activity. Chemosphere 2012, 87, 1273–1280. [Google Scholar] [CrossRef]
- Di Prima, S.; Rodrigo-Comino, J.; Novara, A.; Iovino, M.; Pirastru, M.; Keesstra, S.; Cerdà, A. Soil physical quality of citrus orchards under tillage, herbicide, and organic managements. Pedosphere 2018, 28, 463–477. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liang, T.; Wu, K.; Wang, Y. Shifts in indigenous microbial communities during the anaerobic degradation of pentachlo rophenol in upland and paddy soils from southern China. Environ. Sci. Pollut. Res. Int. 2016, 23, 1–11. [Google Scholar] [CrossRef]
- Han, D.; Yan, D.; Wang, Q.; Fang, W.; Wang, X.; Li, J.; Ouyang, C.B.; Cao, A.C. Effects of soil type, temperature, moisture, appli cation dose, fertilizer and organic amendments on chemical and bio-degradation of dimethyl disulfide in soil. Land Degrad. Dev. 2018, 29, 4282–4290. [Google Scholar] [CrossRef]
- Guo, J.F.; Lu, Y.T.; Sun, J.H. Residue dynamics of fomesafen in peanut and soybean fields. Agroenviron. Prot. 2000, 2, 82–84. (In Chinese) [Google Scholar]
- Hu, H.Y.; Zhou, H.; Zhou, S.X.; Li, Z.J.; Wei, C.J.; Yu, Y.; Hay, A.G. Fomesafen impacts bacterial communities and enzyme activities in the rhizosphere. Environ. Pollut. 2019, 253, 302–311. [Google Scholar] [CrossRef]
- Zhang, Q.M.; Zhu, L.S.; Wang, J.; Xie, H.; Wang, J.H.; Wang, F.H.; Sun, F.X. Effects of fomesafen on soil enzyme activity, microbial population, and bacterial community composition. Environ. Monit. Assess. 2014, 186, 2801–2812. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014 International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014. [Google Scholar]
- Ma, M.; Baskin, C.C.; Yu, K. Wetland drying indirectly influences plant community and seed bank diversity through soil pH. Ecol. Indic. 2017, 80, 186–195. [Google Scholar] [CrossRef]
- Paramasivam, M.; Banerjee, H. Simultaneous determination of flubendiamide its metabolite desiodo flubendiamide residues in cabbage, tomato and pigeon pea by HPLC. Bull. Environ. Contam. Toxicol. 2011, 7, 452–456. [Google Scholar] [CrossRef]
- Che, J.M.; Liu, B.; Zhang, Y.; Hu, G.P.; Huang, Q.L.; Chen, Z.D.; Weng, B.Q. Effect of edible fungus residue on the ecological characteristics of microorganism fatty acids in the rhizosphere of ryegrass. Pratacultural Sci. 2016, 33, 1942–1950. (In Chinese) [Google Scholar]
- Lyu, N.; Yin, F.H.; Chen, Y.; Gao, Z.J.; Liu, Y.; Shi, L. Effects of elevated atmospheric CO2 and nitrogen application on cotton biomass, nitrogen utilization and soil urease activity. Ying Yong Sheng Tai Xue Bao 2015, 26, 3337–3344. (In Chinese) [Google Scholar]
- Zhang, W.; Lin, K.F.; Yang, S.S.; Zhang, M. Enzyme activities in perfluorooctanoic acid (PFOA)-polluted soils. Pedosphere 2013, 23, 120–127. [Google Scholar] [CrossRef]
- Li, Y.L.; Chen, J.X.; Zhou, Y.T.; Yang, Z.; Wang, Y.G.; Zhang, J.Y.; Yue, L. Diversity and distribution of culturable microbial communities of semiarid desert steppe and cultivated land in Northwestern China. J. Agric. Resour. Environ. 2016, 33, 244–252. (In Chinese) [Google Scholar]
- Chen, L.H.; Zheng, J.H.; Shao, X.H. Effects of Trichoderma harzianum T83 on Suaeda salsa L. in coastal saline soil. Ecol. Eng. 2016, 91, 58–64. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, J.; Han, L. Characterization of lignocellulosic compositions’ degradation during chicken manure composting with added biochar by phospholipid fatty acid (PLFA) and correlation analysis. Sci. Total Environ. 2017, 586, 1003–1011. [Google Scholar] [CrossRef]
- Guo, Y.L.; Song, W.F.; Li, M. Environmental behavior of herbicide fomesafen in different soil types. J. Ecol. 2014, 33, 429–432. (In Chinese) [Google Scholar]
- Sebiomo, A.; Ogundero, V.W.; Bankole, S.A. Effect of four herbicide on microbial population, soil organic matter and dehydrogenase activity. Afr. J. Biotechnol. 2011, 10, 770–778. [Google Scholar]
- Zhou, S.X.; Wei, C.J.; Hu, H.Y.; Gao, B.J.; Li, Z.J. Effect of fomesafen on soil microorganism and enzyme activity in soybean rhizosphere and its degradation in rhizosphere. J. Plant Nutr. Fertil. 2018, 24, 203–211. (In Chinese) [Google Scholar]
- Lahl, K.; Unger, C.; Emmerling, C.; Broer, I.; Thiele-Bruhn, S. Response of soil microorganisms and enzyme activities on the decomposition of transgenic cyanophycin producing potatoes during overwintering in soil. Eur. J. Soil Biol. 2012, 53, 1–10. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral nutrition—Current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [Green Version]
- Shentu, J.L.; He, Z.L.; Zeng, Y.Y.; He, S.Y.; Du, S.T.; Shen, D.S. Microbial biomass and PLFA profile changes in rhizosphere of pakchoi (Brassica chinensis L.) as affected by external cadmium loading. Pedosphere 2014, 24, 553–562. [Google Scholar] [CrossRef]
- Piutti, S.; Marchand, A.-L.; Lagacherie, B.; Martin-Laurent, F.; Soulas, G. Effect of cropping cycles and repeated herbicide applications on the degradation of diclofop-methyl, bentazone, diuron, isoproturon and pendimethalin in soil. Pest Manag. Sci. 2002, 58, 303–312. [Google Scholar] [CrossRef]
- Wu, X.H.; Zhang, Y.; Du, P.Q.; Xu, J.; Dong, F.S.; Liu, X.G.; Zheng, Y.Q. Impact of fomesafen on the soil microbial communities in soybean fields in Northeastern China. Ecotoxicol. Environ. Saf. 2018, 148, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Banks, M.L.; Kennedy, A.C.; Kremer, R.J.; Eivazi, F. Soil microbial community response to surfactants and herbicide in two soils. Appl. Soil Ecol. 2014, 74, 12–20. [Google Scholar] [CrossRef]
- Patrnogic, J.; Leclerc, V. The serine protease homolog spheroide is involved in sensing of pathogenic Gram-positive bacteria. PLoS ONE 2017, 12, e0188339. [Google Scholar]





| Treatment | C0(mg·kg−1) | A | k | R2 | t1/2 (d) |
|---|---|---|---|---|---|
| Recommended level | 3.75 | 3.2907 | 0.031 | 0.7248 | 18.14 |
| Twice the recommended level | 7.50 | 4.2745 | 0.012 | 0.7922 | 10.91 |
| Catalase Activity | Acid Phosphatase Activity | Urease Activity | Saccharase Activity | Bacteria | Fungi | Actinomycetes | Fungi/Bacteria | GN/GP | Stress Indicator | Total PLFA | Residue | Predose | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase activity | 1 | ||||||||||||
| Acid phosphatase activity | 0.157 | 1 | |||||||||||
| Urease activity | 0.600 ** | 0.518 ** | 1 | ||||||||||
| Saccharase activity | −0.11 | −0.067 | 0.064 | 1 | |||||||||
| Bacteria | −0.552 ** | −0.247 | −0.436 ** | −0.103 | 1 | ||||||||
| Fungi | 0.215 | 0.183 | 0.435 ** | −0.046 | −0.068 | 1 | |||||||
| Actinomycetes | −0.117 | −0.447 ** | −0.391 ** | −0.448 ** | 0.257 | −0.131 | 1 | ||||||
| Fungi/Bacteria | −0.054 | 0.02 | 0.133 | −0.107 | 0.244 | −0.068 | −0.077 | 1 | |||||
| GN/GP | −0.329 * | −0.197 | −0.096 | −0.014 | 0.306 * | −0.163 | 0.032 | 0.490 ** | 1 | ||||
| Stress Indicator | 0.068 | −0.1 | 0.114 | −0.014 | 0.088 | 0.262 | 0.079 | −0.396 ** | −0.471 ** | 1 | |||
| Total PLFA | −0.209 | 0.132 | −0.047 | −0.025 | 0.579 ** | 0.272 * | −0.016 | 0.317 * | 0.275 * | −0.092 | 1 | ||
| Residue | 0.273 * | −0.001 | −0.075 | −0.198 | −0.321 * | −0.234 | 0.344 * | −0.448 ** | −0.369 ** | 0.123 | −0.362 ** | 1 | |
| Predose | 0.323 * | −0.358** | −0.042 | 0.116 | −0.589 ** | −0.172 | 0.146 | −0.392 ** | −0.119 | −0.065 | −0.665 ** | 0.559 ** | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Wang, Y.; Huang, Y.; Wei, Y.; Yuan, M.; Fu, H.; Zeng, W.; Liu, C. Microbial Community’s Dynamic Response to Fomesafen Usage in Chernozems of Northeast China. Diversity 2021, 13, 340. https://doi.org/10.3390/d13080340
Yang F, Wang Y, Huang Y, Wei Y, Yuan M, Fu H, Zeng W, Liu C. Microbial Community’s Dynamic Response to Fomesafen Usage in Chernozems of Northeast China. Diversity. 2021; 13(8):340. https://doi.org/10.3390/d13080340
Chicago/Turabian StyleYang, Fengshan, Yanbo Wang, Yanan Huang, Yuning Wei, Mingrui Yuan, Haiyan Fu, Weimin Zeng, and Chunguang Liu. 2021. "Microbial Community’s Dynamic Response to Fomesafen Usage in Chernozems of Northeast China" Diversity 13, no. 8: 340. https://doi.org/10.3390/d13080340
APA StyleYang, F., Wang, Y., Huang, Y., Wei, Y., Yuan, M., Fu, H., Zeng, W., & Liu, C. (2021). Microbial Community’s Dynamic Response to Fomesafen Usage in Chernozems of Northeast China. Diversity, 13(8), 340. https://doi.org/10.3390/d13080340