Genetic Diversity and Maternal Lineage of Indo-Pacific Bottlenose Dolphin (Tursiops aduncus) in the Andaman Sea of Thailand
Abstract
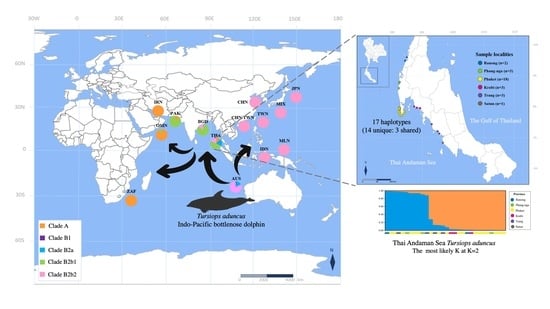
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Microsatellite Amplification and Genotyping
2.3. Control Region (D-Loop) Primer and Amplification
2.4. Data Analysis
2.4.1. Microsatellite Analysis
2.4.2. Mitochondrial DNA Control Region Analysis
2.4.3. Phylogenetic Tree Construction
2.4.4. Bayesian Skyline Plots (BSPs)
3. Results
3.1. Microsatellite Primers and Genetic Diversity
3.2. Microsatellite Forensic Parameter
3.3. Population Structure
3.4. Mitochondrial DNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braulik, G.; Natoli, A.; Kiszka, J.; Parra, G.; Plön, S.; Smith, B. Tursiops aduncus. The IUCN Red List of Threatened Species 2019: E. T41714A50381127. Available online: https://dx.doi.org/10.2305/IUCN.UK.2019-3.RLTS.T41714A50381127.en (accessed on 2 September 2022).
- Department of Marine and Coastal Resources. Indo-Pacific Bottlenose Dolphin, Tursiops aduncus. Central Database System and Data Standard for Marine and Coastal Resources. 2013. Available online: https://km.dmcr.go.th/c_1/s_96/d_4194 (accessed on 25 August 2022).
- Chantrapornsyl, S.; Adulyanukosol, K.; Kittiwathanawong, K. Records of cetaceans in Thailand. Phuket Mar. Biol. Cent. Res. Bull. 1996, 61, 39–63. [Google Scholar]
- Cherdsukjai, P.A.I.; Promjinda, S.; Munprasit, A.; Kittiwattanawong, K. Preliminary Survey of Marine Mammals in Gulf of Thailand. In Survey Results of Fisheries Resources and Marine Environment in the Central Gulf of Thailand by SEAF Survey Boat; South East Asian Fisheries Development Centre: Samut Prakan, Thailand, 2013; pp. 137–141. [Google Scholar]
- Van Bressem, M.-F.; Duignan, P.J.; Banyard, A.; Barbieri, M.; Colegrove, K.M.; De Guise, S.; Di Guardo, G.; Dobson, A.; Domingo, M.; Fauquier, D. Cetacean morbillivirus: Current knowledge and future directions. Viruses 2014, 6, 5145–5181. [Google Scholar] [CrossRef] [Green Version]
- Stephens, N.; Duignan, P.J.; Wang, J.; Bingham, J.; Finn, H.; Lars Bejder, I.; Patterson, A.P.; Holyoake, C. Cetacean morbillivirus in coastal Indo-Pacific bottlenose dolphins, Western Australia. Emerg. Infect. Dis. 2014, 20, 666. [Google Scholar] [CrossRef]
- Kleinertz, S.; Hermosilla, C.; Ziltener, A.; Kreicker, S.; Hirzmann, J.; Abdel-Ghaffar, F.; Taubert, A. Gastrointestinal parasites of free-living Indo-Pacific bottlenose dolphins (Tursiops aduncus) in the Northern Red Sea, Egypt. Parasitol. Res. 2014, 113, 1405–1415. [Google Scholar] [CrossRef]
- Mapunda, E.C.; Othman, O.C.; Akwilapo, L.D.; Bouwman, H.; Mwevura, H. Concentrations of metallic elements in kidney, liver, and lung tissue of Indo-Pacific bottlenose dolphin Tursiops aduncus from coastal waters of Zanzibar, Tanzania. Mar. Pollut. Bull. 2017, 122, 483–487. [Google Scholar] [CrossRef]
- Harino, H.; Ohji, M.; Wattayakorn, G.; Adulyanukosol, K.; Arai, T.; Miyazaki, N. Accumulation of organotin compounds in tissues and organs of dolphins from the coasts of Thailand. Arch. Environ. Contam. Toxicol. 2008, 54, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Bejder, L.; Samuels, A.; Whitehead, H.; Gales, N.; Mann, J.; Connor, R.; Heithaus, M.; Watson-capps, J.; Flaherty, C.; Krützen, M. Decline in relative abundance of bottlenose dolphins exposed to long-term disturbance. Conserv. Biol. 2006, 20, 1791–1798. [Google Scholar] [CrossRef] [Green Version]
- Lusseau, D.; Higham, J. Managing the impacts of dolphin-based tourism through the definition of critical habitats: The case of bottlenose dolphins (Tursiops spp.) in Doubtful Sound, New Zealand. Tour. Manag. 2004, 25, 657–667. [Google Scholar] [CrossRef]
- Natoli, A.; Peddemors, V.M.; Hoelzel, A.R. Population structure of bottlenose dolphins (Tursiops aduncus) impacted by bycatch along the east coast of South Africa. Conserv. Genet. 2008, 9, 627–636. [Google Scholar] [CrossRef]
- Shahid, U.; Khan, M.M.; Nawaz, R.; Razzaq, S.A.; Ayub, S. Bycatch Analysis of Tuna Gillnet Fisheries of Pakistan: An Analysis of Bycatch Data from 2013–2015; IOTC-2016-WPEB12-INF11; World Wide Fund for Nature: Karachi, Pakistan, 2016. [Google Scholar]
- Temple, A.J.; Wambiji, N.; Poonian, C.N.; Jiddawi, N.; Stead, S.M.; Kiszka, J.J.; Berggren, P. Marine megafauna catch in southwestern Indian Ocean small-scale fisheries from landings data. Biol. Conserv. 2019, 230, 113–121. [Google Scholar] [CrossRef]
- Kasuga, M.P.; Varisanga, M.D.; Davenport, T.R.; Jiddawi, N.; Braulik, G.T. Abundance, spatial distribution and threats to Indo-Pacific bottlenose dolphins (Tursiops aduncus) in an Important Marine Mammal Area in Tanzania. West. Indian Ocean J. Mar. Sci. 2022, 21, 1–14. [Google Scholar] [CrossRef]
- Department of National Parks Wildlife and Plant Conservation. Wildlife Conservation and Protection Act, B.E. 2562. Thai Government Gazette. 2019. Available online: http://portal.dnp.go.th/Law (accessed on 20 August 2022).
- Heithaus, M.R.; Frid, A.; Wirsing, A.J.; Worm, B. Predicting ecological consequences of marine top predator declines. Trends Ecol. Evol. 2008, 23, 202–210. [Google Scholar] [CrossRef]
- Wiszniewski, J.; Beheregaray, L.B.; Allen, S.J.; Möller, L.M. Environmental and social influences on the genetic structure of bottlenose dolphins (Tursiops aduncus) in Southeastern Australia. Conserv. Genet. 2010, 11, 1405–1419. [Google Scholar] [CrossRef]
- Yang, G.; Ji, G.; Ren, W.; Zhou, K.; Wei, F. Pattern of genetic variation of bottlenose dolphins in Chinese waters. Raffles Bull Zool 2005, 53, 157–164. [Google Scholar]
- Gray, H.; Nishida, S.; Welch, A.; Moura, A.; Tanabe, S.; Kiani, M.; Culloch, R.; Möller, L.; Natoli, A.; Ponnampalam, L. Cryptic lineage differentiation among Indo-Pacific bottlenose dolphins (Tursiops aduncus) in the northwest Indian Ocean. Mol. Phylogenetics Evol. 2018, 122, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.; Nishida, S.; Yang, W.-C.; Isobe, T.; Tajima, Y.; Hoelzel, A.R. Genetic diversity of bottlenose dolphin (Tursiops sp.) populations in the western North Pacific and the conservation implications. Mar. Biol. 2017, 164, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Piboon, P.; Poommouang, A.; Buddhachat, K.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. The first study of genetic diversity and population structure of Indo-Pacific bottlenose dolphin (Tursiops aduncus) and pantropical spotted dolphin (Stenella attenuata) in the Thai Andaman Sea based on ISSR. Vet. World 2022, 15, 2004–2011. [Google Scholar] [CrossRef]
- Department of Marine and Coastal Resources. Status of Whales and Dolphins in Thai Waters. Central Database System and Data Standard for Marine and Coastal Resources. 2013. Available online: https://km.dmcr.go.th/c_7/d_1812 (accessed on 25 August 2022).
- Hulce, D.; Li, X.; Snyder-Leiby, T.; Liu, C.J. GeneMarker® genotyping software: Tools to increase the statistical power of DNA fragment analysis. J. Biomol. Tech. JBT 2011, 22, S35. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Blair, D.; McMahon, A.; McDonald, B.; Tikel, D.; Waycott, M.; Marsh, H. Pleistocene sea level fluctuations and the phylogeography of the dugong in Australian waters. Mar. Mammal Sci. 2014, 30, 104–121. [Google Scholar] [CrossRef]
- Kriangwanich, W.; Buddhachat, K.; Poommouang, A.; Chomdej, S.; Thitaram, C.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. Feasibility of melting fingerprint obtained from ISSR-HRM curves for marine mammal species identification. PeerJ 2021, 9, e11689. [Google Scholar] [CrossRef] [PubMed]
- Piboon, P.; Poommouang, A.; Buddhachat, K.; Kampuansai, J.; Chomdej, S.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. The Potential Distribution and Maternal Lineage of Two Cetaceans Species (Grampus griseus and Pseudorca crassidens) in the Subfamily Globicephalinae from the Thai Andaman Sea and the Gulf of Thailand. Diversity 2022, 14, 257. [Google Scholar] [CrossRef]
- Piboon, P.; Kriengsakpichit, N.; Poommouang, A.; Buddhachat, K.; Brown, J.L.; Kampuansai, J.; Chomdej, S.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. Relationship of stranded cetaceans in Thai territorial waters to global populations: Mitochondrial DNA diversity of Cuvier’s beaked whale, Indo Pacific finless porpoise, pygmy sperm whale, and dwarf sperm whale. Sci. Prog. 2022, 105, 00368504221103776. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Wang, J. A parsimony estimator of the number of populations from a STRUCTURE-like analysis. Mol. Ecol. Resour. 2019, 19, 970–981. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.; Teslenko, M.; Nylander, J. MrBayes Version 3.2 Manual: Tutorials and Model Summaries. 2011. Available online: http://brahms.biology.rochester.edu/software.html (accessed on 5 August 2022).
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.; Rambaut, A.; Bouckaert, R.; Xie, W. Divergence Dating Tutorial with BEAST 2.0. 2012. Available online: http://beast2.cs.auckland.ac.nz/ (accessed on 5 August 2022).
- Moura, A.E.; Nielsen, S.C.; Vilstrup, J.T.; Moreno-Mayar, J.V.; Gilbert, M.T.P.; Gray, H.W.; Natoli, A.; Möller, L.; Hoelzel, A.R. Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst. Biol. 2013, 62, 865–877. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [Green Version]
- Gray, H.; van Waerebeek, K.; Owen, J.; Collins, T.; Minton, G.; Ponnampalam, L.; Willson, A.; Baldwin, R.; Hoelzel, A.R. Evolutionary drivers of morphological differentiation among three bottlenose dolphin lineages, Tursiops spp. (Delphinidae), in the northwest Indian Ocean utilizing linear and geometric morphometric techniques. Biol. J. Linn. Soc. 2022, 135, 610–629. [Google Scholar] [CrossRef]
- Amaral, A.R.; Smith, B.D.; Mansur, R.M.; Brownell, R.L.; Rosenbaum, H.C. Oceanographic drivers of population differentiation in Indo-Pacific bottlenose (Tursiops aduncus) and humpback (Sousa spp.) dolphins of the northern Bay of Bengal. Conserv. Genet. 2017, 18, 371–381. [Google Scholar] [CrossRef]
- Brown, A.M.; Kopps, A.M.; Allen, S.J.; Bejder, L.; Littleford-Colquhoun, B.; Parra, G.J.; Cagnazzi, D.; Thiele, D.; Palmer, C.; Frere, C.H. Population differentiation and hybridisation of Australian snubfin (Orcaella heinsohni) and Indo-Pacific humpback (Sousa chinensis) dolphins in north-western Australia. PLoS ONE 2014, 9, e101427. [Google Scholar] [CrossRef] [Green Version]
- Guoqing, J.; Guang, Y.; Shan, L.; Kaiya, Z. Variability of the mitochondrial DNA control region of bottlenose dolphins (genus: Tursiops) in Chinese waters. Dong Wu Xue Bao [Acta Zool. Sin.] 2002, 48, 487–493. [Google Scholar]
- Wang, J.; Chou, L.S.; White, B. Mitochondrial DNA analysis of sympatric morphotypes of bottlenose dolphins (genus: Tursiops) in Chinese waters. Mol. Ecol. 1999, 8, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Gridley, T.; Elwen, S.H.; Harris, G.; Moore, D.; Hoelzel, A.; Lampen, F. Hybridization in bottlenose dolphins—A case study of Tursiops aduncus× T. truncatus hybrids and successful backcross hybridization events. PLoS ONE 2018, 13, e0201722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrin, W.F.; Robertson, K.M.; Van Bree, P.J.; Mead, J.G. Cranial description and genetic identity of the holotype specimen of Tursiops aduncus (Ehrenberg, 1832). Mar. Mammal Sci. 2007, 23, 343–357. [Google Scholar] [CrossRef]
- Inamori, D.; Kita, Y.F.; Funasaka, N. External Morphological and Molecular Evidence of Natural Intrageneric Hybridization between Common and Indo-Pacific Bottlenose Dolphins (Tursiops truncatus× T. aduncus) from Japanese Waters. Mammal Study 2021, 47, 133–140. [Google Scholar] [CrossRef]
- Sakai, M.; Kita, Y.F.; Kogi, K.; Shinohara, M.; Morisaka, T.; Shiina, T.; Inoue-Murayama, M. A wild Indo-Pacific bottlenose dolphin adopts a socially and genetically distant neonate. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Yusmalinda, N.L.A.; Anggoro, A.W.; Suhendro, D.M.; Ratha, I.M.J.; Suprapti, D.; Kreb, D.; Cahyani, N.K.D. Species identification of stranded cetaceans in Indonesia revealed by molecular technique. J. Ilmu Dan Teknol. Kelaut. Trop. 2017, 9, 465–474. [Google Scholar] [CrossRef] [Green Version]
- Ansmann, I.C.; Parra, G.J.; Lanyon, J.M.; Seddon, J.M. Fine-scale genetic population structure in a mobile marine mammal: Inshore bottlenose dolphins in M oreton B ay, A ustralia. Mol. Ecol. 2012, 21, 4472–4485. [Google Scholar] [CrossRef]
- Chabanne, D.B.; Allen, S.J.; Sherwin, W.B.; Finn, H.; Krützen, M. Inconsistency between socio-spatial and genetic structure in a coastal dolphin population. Front. Mar. Sci. 2021, 7, 617540. [Google Scholar] [CrossRef]
- Manlik, O.; Chabanne, D.; Daniel, C.; Bejder, L.; Allen, S.J.; Sherwin, W.B. Demography and genetics suggest reversal of dolphin source-sink dynamics, with implications for conservation. Mar. Mammal Sci. 2019, 35, 732–759. [Google Scholar] [CrossRef]
- Möller, L.M.; Beheregaray, L.B. Coastal bottlenose dolphins from southeastern Australia are Tursiops aduncus according to sequences of the mitochondrial DNA control region. Mar. Mammal Sci. 2001, 17, 249–263. [Google Scholar] [CrossRef]
- Möller, L.M.; Wiszniewski, J.; Allen, S.J.; Beheregaray, L.B. Habitat type promotes rapid and extremely localised genetic differentiation in dolphins. Mar. Freshw. Res. 2007, 58, 640–648. [Google Scholar] [CrossRef] [Green Version]
- Oremus, M.; Garrigue, C.; Tezanos-Pinto, G.; Scott Baker, C. Phylogenetic identification and population differentiation of bottlenose dolphins (Tursiops spp.) in Melanesia, as revealed by mitochondrial DNA. Mar. Mammal Sci. 2015, 31, 1035–1056. [Google Scholar] [CrossRef]
- Council, N.R. The Evaluation of Forensic DNA Evidence. 1996. Available online: https://nap.nationalacademies.org/read/5141/chapter/1 (accessed on 11 August 2022).
- Chakraborty, R.; Jin, L. Heterozygote deficiency, population substructure and their implications in DNA fingerprinting. Hum. Genet. 1992, 88, 267–272. [Google Scholar] [CrossRef]
- Haughey, R.; Hunt, T.; Hanf, D.; Rankin, R.W.; Parra, G.J. Photographic capture-recapture analysis reveals a large population of Indo-Pacific bottlenose dolphins (Tursiops aduncus) with low site fidelity off the North West Cape, Western Australia. Front. Mar. Sci. 2020, 6, 781. [Google Scholar] [CrossRef]
- Kiszka, J.; Simon-Bouhet, B.; Gastebois, C.; Pusineri, C.; Ridoux, V. Habitat partitioning and fine scale population structure among insular bottlenose dolphins (Tursiops aduncus) in a tropical lagoon. J. Exp. Mar. Biol. Ecol. 2012, 416, 176–184. [Google Scholar] [CrossRef]
- Natoli, A.; Peddemors, V.M.; Rus Hoelzel, A. Population structure and speciation in the genus Tursiops based on microsatellite and mitochondrial DNA analyses. J. Evol. Biol. 2004, 17, 363–375. [Google Scholar] [CrossRef] [Green Version]
- Särnblad, A.; Danbolt, M.; Dalen, L.; Amir, O.A.; Berggren, P. Phylogenetic placement and population structure of Indo-Pacific bottlenose dolphins (Tursiops aduncus) off Zanzibar, Tanzania, based on mtDNA sequences. Mar. Mammal Sci. 2011, 27, 431–448. [Google Scholar] [CrossRef]
- Xiong, P.; Dudzińska-Nowak, J.; Harff, J.; Xie, X.; Zhang, W.; Chen, H.; Tao, J.; Chen, H.; Miluch, J.; Feldens, P. Modeling paleogeographic scenarios of the last glacial cycle as a base for source-to-sink studies: An example from the northwestern shelf of the South China Sea. J. Asian Earth Sci. 2020, 203, 104542. [Google Scholar] [CrossRef]
- Ponnampalam, L.S. Opportunistic observations on the distribution of cetaceans in the Malaysian South China, Sulu and Sulawesi Seas and an updated checklist of marine mammals in Malaysia. Raffles Bull. Zool. 2012, 60, 221–231. [Google Scholar]
- Lange, I.D.; Schoenig, E.; Khokiattiwong, S. Chapter 22-Thailand. In World Seas: An Environmental Evaluation, 2nd ed.; Sheppard, C., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 491–513. [Google Scholar]
- Möller, L.M.; Beheregaray, L.B. Genetic evidence for sex-biased dispersal in resident bottlenose dolphins (Tursiops aduncus). Mol. Ecol. 2004, 13, 1607–1612. [Google Scholar] [CrossRef]
- Bilgmann, K.; Möller, L.M.; Harcourt, R.G.; Gibbs, S.E.; Beheregaray, L.B. Genetic differentiation in bottlenose dolphins from South Australia: Association with local oceanography and coastal geography. Mar. Ecol. Prog. Ser. 2007, 341, 265–276. [Google Scholar] [CrossRef]
- Vijay, N.; Park, C.; Oh, J.; Jin, S.; Kern, E.; Kim, H.W.; Zhang, J.; Park, J.-K. Population genomic analysis reveals contrasting demographic changes of two closely related dolphin species in the last glacial. Mol. Biol. Evol. 2018, 35, 2026–2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoegh-Guldberg, O.; Bruno, J.F. The impact of climate change on the world’s marine ecosystems. Science 2010, 328, 1523–1528. [Google Scholar] [CrossRef] [PubMed]
- Grant, W.S. Problems and cautions with sequence mismatch analysis and Bayesian skyline plots to infer historical demography. J. Hered. 2015, 106, 333–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heller, R.; Chikhi, L.; Siegismund, H.R. The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS ONE 2013, 8, e62992. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Nishida, S.; Chou, L.S.; Tajima, Y.; Yang, W.C.; Isobe, T.; Yamada, T.K.; Hartman, K.; Hoelzel, A.R. Concordance between genetic diversity and marine biogeography in a highly mobile marine mammal, the Risso’s dolphin. J. Biogeogr. 2018, 45, 2092–2103. [Google Scholar] [CrossRef]
- Escorza-Trevino, S.; Archer, F.I.; Rosales, M.; Lang, A.; Dizon, A.E. Genetic differentiation and intraspecific structure of Eastern Tropical Pacific spotted dolphins, Stenella attenuata, revealed by DNA analyses. Conserv. Genet. 2005, 6, 587–600. [Google Scholar]
- Faria, D.M.; da Silva, J.M., Jr.; Pires Costa, L.; Rezende Paiva, S.; Marino, C.L.; Rollo, M.M., Jr.; Baker, C.S.; Cazerta Farro, A.P. Low mtDNA diversity in a highly differentiated population of spinner dolphins (Stenella longirostris) from the Fernando de Noronha Archipelago, Brazil. PLoS ONE 2020, 15, e0230660. [Google Scholar]
- Farro, A.; Rollo, M.; Silva, J.; Marino, C. Isolation and characterization of microsatellite DNA markers for spinner dolphin (Stenella longirostris). Conser. Genet. 2008, 9, 1319–1321. [Google Scholar]
- Mirimin, L.; Coughlan, J.; Rogan, E.; Cross, T. Tetranucleotide microsatellite loci from the striped dolphin (Stenella coeruleoalba Meyen, 1833). Mol. Ecol. Notes 2006, 6, 493–495. [Google Scholar]






| Locus | Genetic Variabilities | HWE (p-Value) | |||||
|---|---|---|---|---|---|---|---|
| Na | Ne | I | Ho | He | F | ||
| Sl9-69 | 6.00 | 2.25 | 1.14 | 0.55 | 0.55 | 0.02 | ns |
| Sd8 | 14.00 | 9.64 | 2.42 | 0.85 | 0.90 | 0.05 | 0.00 * |
| Sl10-26 | 12.00 | 4.41 | 1.91 | 0.71 | 0.77 | 0.08 | 0.00 * |
| Slo15 | 7.00 | 4.43 | 1.59 | 0.80 | 0.77 | −0.03 | 0.00 * |
| Sco65 | 6.00 | 2.43 | 1.18 | 0.65 | 0.59 | −0.11 | 0.00 * |
| Sl8-49 | 9.00 | 5.08 | 1.83 | 0.68 | 0.80 | 0.15 | 0.00 * |
| Slo4 | 5.00 | 2.53 | 1.18 | 0.46 | 0.60 | 0.24 | 0.00 * |
| EV104 | 6.00 | 3.01 | 1.31 | 0.62 | 0.67 | 0.07 | 0.00 * |
| Slo9 | 7.00 | 4.26 | 1.65 | 0.50 | 0.77 | 0.35 | 0.00 * |
| Sco66 | 7.00 | 4.52 | 1.66 | 0.52 | 0.78 | 0.33 | 0.00 * |
| Slo1 | 7.00 | 3.04 | 1.35 | 0.68 | 0.67 | −0.01 | 0.00 * |
| Mean (SE) | 7.82 (0.84) | 4.15 (0.63) | 1.57 (0.12) | 0.64 (0.04) | 0.72 (0.03) | 0.10 (0.04) | |
| Location | Number of Haplotypes | Haplotype Diversity (h) | Nucleotide Diversity (π) | Reference |
|---|---|---|---|---|
| THA | 17 | 0.926 ± 0.027 | 0.045 ± 0.002 | [45], this study |
| BGD | 7 | 1.000 ± 0.076 | 0.021 ± 0.009 | [46,47] |
| OMN | 9 | 0.624 ± 0.058 | 0.011 ± 0.003 | [45] |
| PAK | 6 | 0.889 ± 0.091 | 0.018 ± 0.010 | [45] |
| CHN | 15 | 0.932 ± 0.031 | 0.020 ± 0.001 | [46,48] |
| MIX | 18 | 1.000 ± 0.019 | 0.020 ± 0.001 | [21] |
| IDN | 3 | 1.000 ± 0.272 | 0.272 ± 0.010 | [49] |
| CHN/TWN | 5 | 1.000 ± 0.126 | 0.019 ± 0.003 | [49] |
| TWN | 5 | 1.000 ± 0.126 | 0.025 ± 0.004 | [49] |
| ZAF | 5 | 0.786 ± 0.151 | 0.007 ± 0.002 | [12,50,51] |
| JPN | 3 | 1.000 ± 0.272 | 0.025 ± 0.007 | [52,53,54] |
| AUS | 40 | 0.989 ± 0.006 | 0.031 ± 0.002 | [47,55,56,57,58,59] |
| MLN | 9 | 1.000 ± 0.052 | 0.013 ± 0.002 | [60] |
| IRN | 1 | 0.000 | 0.000 | [45] |
| Overall | 109 | 0.970 ± 0.006 | 0.042 ± 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasitwiset, W.; Wongfu, C.; Poommouang, A.; Buddhachat, K.; Brown, J.L.; Chomdej, S.; Kampuansai, J.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K.; et al. Genetic Diversity and Maternal Lineage of Indo-Pacific Bottlenose Dolphin (Tursiops aduncus) in the Andaman Sea of Thailand. Diversity 2022, 14, 1093. https://doi.org/10.3390/d14121093
Prasitwiset W, Wongfu C, Poommouang A, Buddhachat K, Brown JL, Chomdej S, Kampuansai J, Kaewmong P, Kittiwattanawong K, Nganvongpanit K, et al. Genetic Diversity and Maternal Lineage of Indo-Pacific Bottlenose Dolphin (Tursiops aduncus) in the Andaman Sea of Thailand. Diversity. 2022; 14(12):1093. https://doi.org/10.3390/d14121093
Chicago/Turabian StylePrasitwiset, Wareerat, Chutima Wongfu, Anocha Poommouang, Kittisak Buddhachat, Janine L. Brown, Siriwadee Chomdej, Jatupol Kampuansai, Patcharaporn Kaewmong, Kongkiat Kittiwattanawong, Korakot Nganvongpanit, and et al. 2022. "Genetic Diversity and Maternal Lineage of Indo-Pacific Bottlenose Dolphin (Tursiops aduncus) in the Andaman Sea of Thailand" Diversity 14, no. 12: 1093. https://doi.org/10.3390/d14121093
APA StylePrasitwiset, W., Wongfu, C., Poommouang, A., Buddhachat, K., Brown, J. L., Chomdej, S., Kampuansai, J., Kaewmong, P., Kittiwattanawong, K., Nganvongpanit, K., & Piboon, P. (2022). Genetic Diversity and Maternal Lineage of Indo-Pacific Bottlenose Dolphin (Tursiops aduncus) in the Andaman Sea of Thailand. Diversity, 14(12), 1093. https://doi.org/10.3390/d14121093