Mitochondrial Genetic Diversity and Structure of the Langur Population in a Complex Landscape of the Nepal Himalaya
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Fecal Sampling and DNA Extraction
2.3. PCR Amplification and DNA Sequencing
2.4. Data Analysis
2.4.1. DNA Polymorphism and Genetic Diversity
2.4.2. Inter-Haplotypic Relationships
2.4.3. Isolation by Distance
2.4.4. Isolation by Resistance
3. Results
3.1. Genetic Diversity
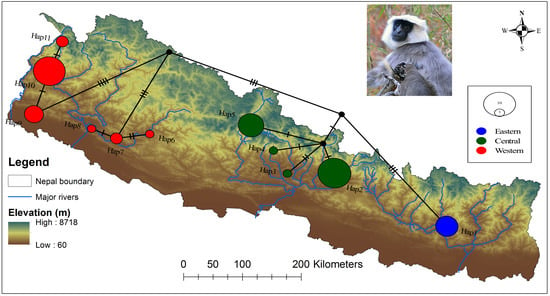
3.2. Population Genetic Structure
3.3. Isolation by Distance and Isolation by Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shrestha, U.B.; Gautam, S.; Bawa, K.S. Widespread climate change in the Himalayas and associated changes in local ecosystems. PLoS ONE 2012, 7, e36741. [Google Scholar] [CrossRef] [Green Version]
- Owen, L.A.; Caffee, M.W.; Finkel, R.C.; Seong, Y.B. Quaternary glaciation of the Himalayan-Tibetan orogen. J. Quat. Sci. 2008, 23, 513–531. [Google Scholar] [CrossRef] [Green Version]
- Pisano, J.; Condamine, F.L.; Lebedev, V.; Bannikova, A.; Quéré, J.-P.; Shenbrot, G.I.; Pagès, M.; Michaux, J.R.; Riddle, B. Out of Himalaya: The impact of past Asian environmental changes on the evolutionary and biogeographical history of Dipodoidea (Rodentia). J. Biogeogr. 2015, 42, 856–870. [Google Scholar] [CrossRef]
- Pandit, M.K.; Manish, K.; Koh, L.P. Dancing on the Roof of the World: Ecological Transformation of the Himalayan Landscape. Bioscience 2014, 64, 980–992. [Google Scholar] [CrossRef] [Green Version]
- Jackson, N.D.; Fahrig, L. Habitat amount, not habitat configuration, best predicts population genetic structure in fragmented landscapes. Landsc. Ecol. 2015, 31, 951–968. [Google Scholar] [CrossRef]
- Khanal, L.; Chalise, M.K.; Wan, T.; Jiang, X.L. Riverine barrier effects on population genetic structure of the Hanuman langur (Semnopithecus entellus) in the Nepal Himalaya. BMC Evol. Biol. 2018, 18, 159. [Google Scholar] [CrossRef] [PubMed]
- Manel, S.; Schwarts, M.K.; Luikart, G.; Taberlet, P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Smouse, P.E.; Peakall, R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 1999, 82, 561–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanal, L.; Chalise, M.K.; Jiang, X. Ecological niche modelling of Himalayan Langur (Semnopithecus entellus) in southern flank of the Himalaya. J. Inst. Sci. Technol. 2018, 23, 1–9. [Google Scholar] [CrossRef]
- Bishop, N.H. Himalayan langurs: Temperate colobines. J. Hum. Evol. 1979, 8, 251–281. [Google Scholar] [CrossRef]
- Curtin, R.A. Range use of gray langurs in highland Nepal. Folia Primatol. 1982, 38, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Borries, C.; Sommer, V.; Srivastava, A. Dominance, age, and reproductive success in free-ranging female Hanuman langurs (Presbytis entellus). Int. J. Primatol. 1991, 12, 231–257. [Google Scholar] [CrossRef]
- Ostner, J.; Chalise, M.K.; Koenig, A.; Launhardt, K.; Nikolei, J.; Podzuweit, D.; Borries, C. What Hanuman langur males know about female reproductive status. Am. J. Primatol. 2006, 68, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Borries, C.; Perlman, R.F.; Koenig, A. Characteristics of alpha males in Nepal gray langurs. Am. J. Primatol. 2017, 79, e22437. [Google Scholar] [CrossRef]
- Perlman, R.F.; Borries, C.; Koenig, A. Dominance relationships in male Nepal gray langurs (Semnopithecus schistaceus). Am. J. Phys. Anthropol. 2016, 160, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Chalise, M.K. Fragmented primate population of Nepal. In Primates in Fragments: Complexity and Resilience; Marsh, L.K., Chapman, C.A., Eds.; Springer Science+Business Media: New York, NY, USA, 2013; pp. 329–356. [Google Scholar] [CrossRef]
- Ale, P.B.; Kandel, K.; Ghimire, T.R.; Huettmann, F.; Regmi, G.R. Persistent evidence for a dramatic decline in langurs (Semnopithecus spp.) in Nepal and elsewhere: Science data and personal experiences converge on a landscape-scale. In Hindu Kush-Himalaya Watersheds Downhill: Landscape Ecology and Conservation Perspectives; Regmi, G.R., Huettmann, F., Eds.; Springer Nature: Cham, Switzerland, 2020; pp. 663–676. [Google Scholar]
- Bagaria, P.; Sharma, L.K.; Joshi, B.D.; Kumar, H.; Mukherjee, T.; Thakur, M.; Chandra, K. West to east shift in range predicted for Himalayan Langur in climate change scenario. Glob. Ecol. Conserv. 2020, 22, e00926. [Google Scholar] [CrossRef]
- Di Fiore, A. Molecular genetic approaches to the study of primate behavior, social organization, and reproduction. Am. J. Phys. Anthropol. 2003, 122, 62–99. [Google Scholar] [CrossRef]
- Chalise, M.K.; Karki, J.B.; Ghimire, M.K. Status in Nepal: Non-human primates. In Special Issue on the Occasion of 10th Wildlife Week, 2062; Department of National Parks & Wildlife Conservation: Kathmandu, Nepal, 2005; pp. 19–26. [Google Scholar]
- Singh, M.; Kumar, A.; Kumara, H.N.; Ahuja, V. Semnopithecus hector. The IUCN Red List of Threatened Species 2020: E.T39837A17942651. Available online: https://www.iucnredlist.org/species/39837/17942651 (accessed on 18 August 2021).
- Singh, M.; Kumara, H.N.; Long, Y.; Chetry, D.; Kumar, A. Semnopithecus schistaceus. The IUCN Red List of Threatened Species 2020: E.T39840A17942792. Available online: https://www.iucnredlist.org/species/39840/17942792 (accessed on 18 August 2021).
- Kumar, A.; Singh, M.; Anandam, M.; Ahuja, V.; Kumara, H.N.; Molur, S. Semnopithecus ajax. The IUCN Red List of Threatened Species 2020: E.T39833A17943210. Available online: https://www.iucnredlist.org/species/39833/17943210 (accessed on 3 October 2021).
- Wang, X.P.; Yu, L.; Roos, C.; Ting, N.; Chen, C.P.; Wang, J.; Zhang, Y.P. Phylogenetic Relationships among the Colobine Monkeys Revisited: New Insights from Analyses of Complete mt Genomes and 44 Nuclear Non-Coding Markers. PLoS ONE 2012, 7, e36274. [Google Scholar] [CrossRef] [Green Version]
- Arekar, K.; Sathyakumar, S.; Karanth, K.P. Integrative taxonomy confirms the species status of the Himalayan langurs, Semnopithecus schistaceus Hodgson, 1840. J. Zool. Syst. Evol. Res. 2020, 59, 543–556. [Google Scholar] [CrossRef]
- Sharma, C.K. Physiography. In Nepal- Nature’s Paradise; Majupuria, T.C., Majupiria, R.K., Eds.; Hari Devi: Gwalior, India, 1999; pp. 4–8. [Google Scholar]
- Carson, B.; Shah, P.B.; Maharjan, P.B. Land Systems Report—The Soil Landscapes of Nepal; Kenting Earth Sciences Limited: Kathmandu, Nepal, 1986; p. 140. [Google Scholar]
- Asahi, K. Equilibrium-line altitudes of the present and Last Glacial Maximum in the eastern Nepal Himalayas and their implications for SW monsoon climate. Quat. Int. 2010, 212, 26–34. [Google Scholar] [CrossRef]
- White, P.S.; Densmore, L.D., III. Mitochondrial DNA Isolation; Oxford University Press: Oxford, UK, 1992; pp. 29–58. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree v1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 April 2019).
- Excoffier, L.; Lischer, H.E. Arlequin suite version 3.5: A New series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Ersts, P.J.; Geographic Distance Matrix Generator (v.1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation. Available online: http://biodiversityinformatics.amnh.org/open_source/gdmg (accessed on 15 June 2020).
- Motulsky, H.J. Analyzing Data with GraphPad Prism; GraphPad Software Inc.: San Diego, CA, USA, 1999. [Google Scholar]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudık, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Adams, R.V.; Lazerte, S.E.; Otter, K.A.; Burg, T.M. Influence of landscape features on the microgeographic genetic structure of a resident songbird. Heredity 2016, 117, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McRae, B.H.; Beier, P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. USA 2007, 104, 19885–19890. [Google Scholar] [CrossRef] [Green Version]
- McRae, B.; Shah, V.; Mohapatra, T.T. Circuitscape 4 User Guide. 2011. Available online: http://www.circuitscape.org (accessed on 8 September 2021).
- Ram, M.S.; Kittur, S.M.; Biswas, J.; Nag, S.; Shil, J.; Umapathy, G. Genetic diversity and structure among isolated populations of the endangered Gees Golden Langur in Assam, India. PLoS ONE 2016, 11, e0161866. [Google Scholar] [CrossRef]
- Wang, W.; Qiao, Y.; Pan, W.; Yao, M. Low genetic diversity and strong geographical structure of the critically endangered White-Headed Langur (Trachypithecus leucocephalus) inferred from mitochondrial DNA control region sequences. PLoS ONE 2015, 10, e0129782. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Yang, Y.; Cui, D.; Fickenscher, G.; Zinner, D.; Roos, C.; Brameier, M. Population genetic structure of Guizhou snub-nosed monkeys (Rhinopithecus brelichi) as inferred from mitochondrial control region sequences, and comparison with R. roxellana and R. bieti. Am. J. Phys. Anthropol. 2012, 147, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nabholz, B.; Glemin, S.; Galtier, N. The erratic mitochondrial clock: Variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol. Biol. 2009, 9, 54. [Google Scholar] [CrossRef] [Green Version]
- Pesole, G.; Gissi, C.; Chirico, A.D.; Saccone, C. Nucleotide substitution rate of Mammalian mitochondrial genomes. J. Mol. Evol. 1999, 48, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Guschanski, K.; Caillaud, D.; Robbins, M.M.; Vigilant, L. Females shape the genetic structure of a gorilla population. Curr. Biol. 2008, 18, 1809–1814. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, Y.; Takemoto, H.; Higuchi, S.; Sakamaki, T.; Hart, J.A.; Hart, T.B.; Tokuyama, N.; Reinartz, G.E.; Guislain, P.; Dupain, J.; et al. Genetic structure of wild bonobo populations: Diversity of mitochondrial DNA and geographical distribution. PLoS ONE 2013, 8, e59660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boubli, J.P.; Ribas, C.; Lynch Alfaro, J.W.; Alfaro, M.E.; da Silva, M.N.; Pinho, G.M.; Farias, I.P. Spatial and temporal patterns of diversification on the Amazon: A test of the riverine hypothesis for all diurnal primates of Rio Negro and Rio Branco in Brazil. Mol. Phylogen. Evol. 2015, 82, 400–412. [Google Scholar] [CrossRef]
- Merces, M.P.; Lynch Alfaro, J.W.; Ferreira, W.A.; Harada, M.L.; Silva Junior, J.S. Morphology and mitochondrial phylogenetics reveal that the Amazon River separates two eastern squirrel monkey species: Saimiri sciureus and S. collinsi. Mol. Phylogen. Evol. 2015, 82, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ren, B.; Wu, R.; Zhao, L.; Hao, Y.; Wang, B.; Wei, F.; Long, Y.; Li, M. The effect of landscape features on population genetic structure in Yunnan snub-nosed monkeys (Rhinopithecus bieti) implies an anthropogenic genetic discontinuity. Mol. Ecol. 2009, 18, 3831–3846. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yao, M. Fine-scale genetic structure analyses reveal dispersal patterns in a critically endangered primate, Trachypithecus leucocephalus. Am. J. Primatol. 2017, 79, e22635. [Google Scholar] [CrossRef] [PubMed]
- Brandon-Jones, D. A taxonmic revision of the langurs and leaf monkeys (Primates: Colobinae) of South Asia. Zoos’ Print J. 2004, 19, 1552–1594. [Google Scholar] [CrossRef]





| Region | Area | Latitude (N) | Longitude (E) | ASL (m) | Troop Size | # Samples | # Sequences |
|---|---|---|---|---|---|---|---|
| Eastern | Mulghat | 26°56′07.21″ | 87°19′56.14″ | 280 | 24 | 6 | 3 |
| Tamku | 27°27′34.64″ | 87°18′58.79″ | 556 | 23 | 5 | 4 | |
| Central | Rishing | 28°10′21.28″ | 85°21′01.94″ | 1950 | 39 | 6 | 5 |
| Khanjim | 28°10′05.63″ | 85°21′40.75″ | 2508 | 24 | 4 | 2 | |
| Syafrubeshi | 28°09′35.82″ | 85°20′53.77″ | 1477 | 9 | 3 | 2 | |
| Khalte | 27°51′03.80″ | 84°59′29.30″ | 689 | 24 | 6 | 4 | |
| Baseri | 27°50′28.15″ | 84°45′59.61″ | 382 | 19 | 3 | 1 | |
| Saattale | 28°27′05.73″ | 84°22′33.25″ | 1533 | 32 | 4 | 1 | |
| Kushma | 28°13′37.42″ | 83°40′26.34″ | 877 | 14 | 5 | 4 | |
| Dhairing | 28°13′19.53″ | 83°35′43.53″ | 1080 | 7 | 3 | 2 | |
| Balewa | 28°11′30.56″ | 83°39′07.74″ | 685 | 21 | 8 | 3 | |
| Western | Reshunga | 28°04′24.54″ | 83°15′42.14″ | 1748 | 13 | 3 | 1 |
| Banke * | 28°35′09.47″ | 81°17′06.51″ | 225 | 12 | 2 | 0 * | |
| Chisapani | 28°37′59.05″ | 81°16′57.72″ | 212 | 25 | 3 | 2 | |
| Suklaphanta NP | 28°50′09.90″ | 80°09′02.28″ | 180 | 11 | 5 | 5 | |
| Okhreni | 29°52′43.80″ | 80°54′58.60″ | 2513 | 24 | 9 | 4 | |
| Dhaumula | 29°53′48.90″ | 80°56′11.12″ | 3328 | 12 | 4 | 4 | |
| Dhaulo Odhar | 29°56′41.06″ | 80°56′27.78″ | 3798 | 21 | 8 | 5 | |
| Total | 354 | 87 | 52 | ||||
| Loci | Region | Eastern | Central | Western |
|---|---|---|---|---|
| Number of Sequences (Troops) | 7 (2) | 24 (9) | 21 (6) | |
| COI | PS (PI sites) | 0 (0) | 5 (2) | 10 (8) |
| #H | 1 | 4 | 6 | |
| HD ± SD | 0.000 | 0.587 ± 0.064 | 0.600 ± 0.101 | |
| π ± SD | 0.000 | 0.0019 ± 0.0010 | 0.0030 ± 0.0018 | |
| COI + CR + CYTB | PS (PI sites) | 6 (3) | 92 (49) | 92 (80) |
| H | 6 (Hap1–Hap6) | 20 (Hap7–Hap26) | 9 (Hap27–Hap35) | |
| HD ± SD | 0.952 ± 0.096 | 0.982 ± 0.018 | 0.781 ± 0.07 | |
| π ± SD | 0.00085 ± 0.00001 | 0.0085 ± 0.0028 | 0.0090 ± 0.0030 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khanal, L.; Chalise, M.K.; Jiang, X.-L.; Kyes, R.C. Mitochondrial Genetic Diversity and Structure of the Langur Population in a Complex Landscape of the Nepal Himalaya. Diversity 2022, 14, 69. https://doi.org/10.3390/d14020069
Khanal L, Chalise MK, Jiang X-L, Kyes RC. Mitochondrial Genetic Diversity and Structure of the Langur Population in a Complex Landscape of the Nepal Himalaya. Diversity. 2022; 14(2):69. https://doi.org/10.3390/d14020069
Chicago/Turabian StyleKhanal, Laxman, Mukesh Kumar Chalise, Xue-Long Jiang, and Randall C. Kyes. 2022. "Mitochondrial Genetic Diversity and Structure of the Langur Population in a Complex Landscape of the Nepal Himalaya" Diversity 14, no. 2: 69. https://doi.org/10.3390/d14020069
APA StyleKhanal, L., Chalise, M. K., Jiang, X. -L., & Kyes, R. C. (2022). Mitochondrial Genetic Diversity and Structure of the Langur Population in a Complex Landscape of the Nepal Himalaya. Diversity, 14(2), 69. https://doi.org/10.3390/d14020069