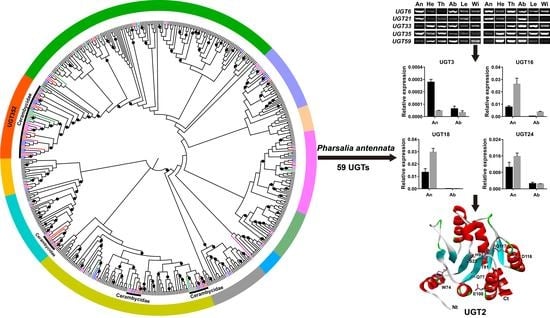
Identification and Characterization of UDP-Glycosyltransferase Genes in a Cerambycid Beetle, Pharsalia antennata Gahan, 1894 (Coleoptera: Cerambycidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Tissue Collection
2.2. RNA Isolation and cDNA Synthesis
2.3. Gene Identification
2.4. Sequence Analysis
2.5. Phylogenetic Tree Construction
2.6. Expression Profiling Analysis
2.7. Homology Modeling of P. antennata UGT2
3. Results
3.1. Identification of Candidate UGT Genes in P. antennata
3.2. Sequence Characteristics of P. antennata UGTs
3.3. Phylogenetic Analysis of Coleopteran UGTs
3.4. Sex- and Tissue-Specific Expression Profile of P. antennata UGTs
3.5. Candidate P. antennata UGTs Involved in Olfaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bruinsma, M.; Dicke, M. Herbivore-Induced Indirect Defense: From Induction Mechanisms to Community Ecology. In Induced Plant Resistance to Herbivory; Schaller, A., Ed.; Springer: Dordrecht, The Netherland, 2008; pp. 31–60. [Google Scholar]
- Despres, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Ferry, N.; Edwards, M.G.; Gatehouse, J.A.; Gatehouse, A.M. Plant-insect interactions: Molecular approaches to insect resistance. Curr. Opin. Biotechnol. 2004, 15, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W. Vertebrate UDP-glucuronosyltransferases: Functional and evolutionary aspects. Biochem. Pharmacol. 2003, 66, 691–696. [Google Scholar] [CrossRef]
- Bock, K.W. The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: Animal-plant arms-race and co-evolution. Biochem. Pharmacol. 2016, 99, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef]
- Ahn, S.J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Chertemps, T.; Maïbèche, M.; Marygold, S.J.; Van Leeuwen, T. Editorial: Invertebrate UDP-Glycosyltransferases: Nomenclature, Diversity and Functions. Front. Physiol. 2021, 12, 748290. [Google Scholar] [CrossRef]
- Daimon, T.; Hirayama, C.; Kanai, M.; Ruike, Y.; Meng, Y.; Kosegawa, E.; Nakamura, M.; Tsujimoto, G.; Katsuma, S.; Shimada, T. The silkworm Green b locus encodes a quercetin 5-O-glucosyltransferase that produces green cocoons with UV-shielding properties. Proc. Natl. Acad. Sci. USA 2010, 107, 11471–11476. [Google Scholar] [CrossRef] [Green Version]
- Luque, T.; Okano, K.; O’Reilly, D.R. Characterization of a novel silkworm (Bombyx mori) phenol UDP-glucosyltransferase. Eur. J. Biochem. 2002, 269, 819–825. [Google Scholar] [CrossRef]
- Krempl, C.; Sporer, T.; Reichelt, M.; Ahn, S.J.; Heidel-Fischer, H.; Vogel, H.; Heckel, D.G.; Joußen, N. Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 71, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Israni, B.; Wouters, F.C.; Luck, K.; Seibel, E.; Ahn, S.J.; Paetz, C.; Reinert, M.; Vogel, H.; Erb, M.; Heckel, D.G.; et al. The fall armyworm Spodoptera frugiperda utilizes specific UDP-glycosyltransferases to inactivate maize defensive benzoxazinoids. Front. Physiol. 2020, 11, 604754. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.N.; Zhao, Y.J.; Zhu, J.Y.; Liu, N.Y. Antennal UDP-glycosyltransferase genes in the coffee white stemborer, Xylotrechus quadripes. J. Asia-Pac. Entomol. 2019, 22, 1145–1153. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Zhou, J.J.; Yi, J.K.; Pan, Y.; Wang, J.; Zhang, X.X.; Wang, J.X.; Yang, S.; Xi, J.H. Identification and tissue expression profiling of candidate UDP-glycosyltransferase genes expressed in Holotrichia parallela motschulsky antennae. B Entomol. Res. 2018, 108, 807–816. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Ma, J.F.; Xu, L.; Dong, Z.P.; Xu, J.W.; Li, M.Y.; Zhu, X.Y. Identification and expression patterns of UDP-glycosyltransferase (UGT) genes from insect pest Athetis lepigone (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 2017, 20, 253–259. [Google Scholar] [CrossRef]
- Younus, F.; Chertemps, T.; Pearce, S.L.; Pandey, G.; Bozzolan, F.; Coppin, C.W.; Russell, R.J.; Maibeche-Coisne, M.; Oakeshott, J.G. Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 2014, 53, 30–43. [Google Scholar] [CrossRef] [Green Version]
- Bozzolan, F.; Siaussat, D.; Maria, A.; Durand, N.; Pottier, M.A.; Chertemps, T.; Maibeche-Coisne, M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 2014, 23, 539–549. [Google Scholar] [CrossRef]
- Wang, Q.; Hasan, G.; Pikielny, C.W. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 1999, 274, 10309–10315. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Xia, J.; Shang, Q.; Song, D.; Gao, X. UDP-glucosyltransferases potentially contribute to imidacloprid resistance in Aphis gossypii glover based on transcriptomic and proteomic analyses. Pestic. Biochem. Physiol. 2019, 159, 98–106. [Google Scholar] [CrossRef]
- Pan, Y.; Wen, S.; Chen, X.; Gao, X.; Zeng, X.; Liu, X.; Tian, F.; Shang, Q. UDP-glycosyltransferases contribute to spirotetramat resistance in Aphis gossypii Glover. Pestic. Biochem. Physiol. 2020, 166, 104565. [Google Scholar] [CrossRef]
- Yang, N.; Xie, W.; Yang, X.; Wang, S.; Wu, Q.; Li, R.; Pan, H.; Liu, B.; Shi, X.; Fang, Y.; et al. Transcriptomic and proteomic responses of sweetpotato whitefly, Bemisia tabaci, to thiamethoxam. PLoS ONE 2013, 8, e61820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, F.; Wang, Z.; Li, C.; Liu, J.; Zeng, X. UDP-Glycosyltransferases are involved in imidacloprid resistance in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Lividae). Pestic. Biochem. Physiol. 2019, 154, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fu, W.B.; Si, F.L.; Yan, Z.T.; Zhang, Y.J.; He, Q.Y.; Chen, B. UDP-glycosyltransferase genes and their association and mutations associated with pyrethroid resistance in Anopheles sinensis (Diptera: Culicidae). Malar. J. 2019, 18, 62. [Google Scholar] [CrossRef] [Green Version]
- Pedra, J.H.F.; McIntyre, L.M.; Scharf, M.E.; Pittendrigh, B.R. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 7034–7039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vontas, J.; Blass, C.; Koutsos, A.C.; David, J.P.; Kafatos, F.C.; Louis, C.; Hemingway, J.; Christophides, G.K.; Ranson, H. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol. Biol. 2005, 14, 509–521. [Google Scholar] [CrossRef]
- Li, X.; Zhu, B.; Gao, X.; Liang, P. Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 2017, 73, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 2016, 17, 227. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Q.; Yin, N.N.; Zhao, N.; Liu, N.Y. Identification and expression characterization of UDP-glucosyltransferase genes in Rhaphuma horsfieldi. Chin. J. Appl. Entomol. 2020, 57, 898–910. [Google Scholar]
- Wu, Z.; Bin, S.; He, H.; Wang, Z.; Li, M.; Lin, J. Differential expression analysis of chemoreception genes in the striped flea beetle Phyllotreta striolata using a transcriptomic approach. PLoS ONE 2016, 11, e0153067. [Google Scholar] [CrossRef] [Green Version]
- Kaplanoglu, E.; Chapman, P.; Scott, I.M.; Donly, C. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Sci. Rep. 2017, 7, 1762. [Google Scholar] [CrossRef] [Green Version]
- Yin, N.N.; Zhao, Y.J.; Zhao, N.; Liu, N.Y. Morphological characteristics of Pharsalia antennata and its sensilla ultrastructure. Plant. Protection. 2020, 46, 30–40. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nicholas, K.B.; Nicholas, H.B.; Deerfield, D.W. GeneDoc: Analysis and visualization of genetic variation. EMBNEW News 1997, 4, 14. [Google Scholar]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010, 5, e9490. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [Green Version]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 2002, 32, 1372–1374, 1376, 1378–1379. [Google Scholar]
- Miley, M.J.; Zielinska, A.K.; Keenan, J.E.; Bratton, S.M.; Radominska-Pandya, A.; Redinbo, M.R. Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J. Mol. Biol. 2007, 369, 498–511. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allison, J.D.; Borden, J.H.; Seybold, S.J. A review of the chemical ecology of the Cerambycidae (Coleoptera). Chemoecology 2004, 14, 123–150. [Google Scholar] [CrossRef]
- Trowbridge, A.M. Evolutionary ecology of chemically mediated plant-insect interactions. In Ecology and the Environment; Monson, R.K., Ed.; Springer: New York, NY, USA, 2014; pp. 1–28. [Google Scholar]
- Nagare, M.; Ayachit, M.; Agnihotri, A.; Schwab, W.; Joshi, R. Glycosyltransferases: The multifaceted enzymatic regulator in insects. Insect Mol. Biol. 2021, 30, 123–137. [Google Scholar] [CrossRef]
- Wang, X.; Fang, X.; Yang, P.; Jiang, X.; Jiang, F.; Zhao, D.; Li, B.; Cui, F.; Wei, J.; Ma, C.; et al. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014, 5, 2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Zhang, S.H.; Ren, M.M.; Tian, X.R.; Wei, Q.; Mburu, D.K.; Su, J.Y. The expression of Spodoptera exigua P450 and UGT genes: Tissue specificity and response to insecticides. Insect Sci. 2019, 26, 199–216. [Google Scholar]
- Li, X.; Shi, H.; Gao, X.; Liang, P. Characterization of UDP-glucuronosyltransferase genes and their possible roles in multi-insecticide resistance in Plutella xylostella (L.). Pest. Manag. Sci. 2018, 74, 695–704. [Google Scholar] [CrossRef]
- Chen, X.; Tang, C.; Ma, K.; Xia, J.; Song, D.; Gao, X.W. Overexpression of UDP-glycosyltransferase potentially involved in insecticide resistance in Aphis gossypii Glover collected from Bt cotton fields in China. Pest. Manag. Sci. 2020, 76, 1371–1377. [Google Scholar] [CrossRef]
- Hopkins, T.L.a.; Kramer, K.J. Insect Cuticle Sclerotization. Annu.Rev. Entomol. 1992, 37, 273–302. [Google Scholar] [CrossRef]
- Kramer, K.J.; Hopkins, T.L. Tyrosine metabolism for insect cuticle tanning. Arch. Insect Biochem. Physiol. 1987, 6, 279–301. [Google Scholar]
- Fraichard, S.; Legendre, A.; Lucas, P.; Chauvel, I.; Faure, P.; Neiers, F.; Artur, Y.; Briand, L.; Ferveur, J.F.; Heydel, J.M. Modulation of sex pheromone discrimination by a UDP-glycosyltransferase in Drosophila melanogaster. Genes 2020, 11, 237. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Zhang, Y.F.; Hong, D.Y.; Wang, J.; Wang, X.L.; Zuo, L.H.; Tang, X.F.; Xu, W.-M.; He, M. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genomics 2017, 18, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiesen, B.; Krug, E.; Fiedler, K.; Wray, V.; Proksch, P. Sequestration of host-plant-derived flavonoids by lycaenid butterfly Polyommatus icarus. J. Chem. Ecol. 1994, 20, 2523–2538. [Google Scholar] [CrossRef] [PubMed]
- Königer, A.; Grath, S. Transcriptome analysis reveals candidate genes for cold tolerance in Drosophila ananassae. Genes 2018, 9, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimbrell, D.A.; Hice, C.; Bolduc, C.; Kleinhesselink, K.; Beckingham, K. The Dorothy enhancer has tinman binding sites and drives hopscotch-induced tumor formation. Genesis 2002, 34, 23–28. [Google Scholar] [CrossRef]
- Zhou, Z.; Rodriguez, A.; Wu, C.Y.; Kimbrell, D.A. Drosophila cellular immune system: Dorothy encodes a UDP glycosyltransferase. Adv. Exp. Med. Biol. 2001, 484, 251–263. [Google Scholar]
- Huang, F.F.; Chai, C.L.; Zhang, Z.; Liu, Z.H.; Dai, F.Y.; Lu, C.; Xiang, Z.H. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 2008, 9, 563. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.-J.; Marygold, S.J. The UDP-glycosyltransferase family in Drosophila melanogaster: Nomenclature update, gene expression and phylogenetic analysis. Front. Physiol. 2021, 12, 648481. [Google Scholar] [CrossRef]
- Durand, N.; Carot-Sans, G.; Bozzolan, F.; Rosell, G.; Siaussat, D.; Debernard, S.; Chertemps, T.; Maibeche-Coisne, M. Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS ONE 2011, 6, e29147. [Google Scholar] [CrossRef]
- Godoy, R.; Machuca, J.; Venthur, H.; Quiroz, A.; Mutis, A. An overview of antennal esterases in Lepidoptera. Front. Physiol. 2021, 12, 643281. [Google Scholar] [CrossRef]
- Xu, W.; Liao, Y. Identification and characterization of aldehyde oxidases (AOXs) in the cotton bollworm. Naturwissenschaften 2017, 104, 94. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Tang, Y.; Wang, Q.; Shi, H.; Yin, J.; Li, C. Identification and characterization of an antennae-specific glutathione S-transferase from the Indian meal moth. Front. Physiol. 2021, 12, 727619. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R. Evolution of insect P450. Biochem. Soc. Trans. 2006, 34, 1252–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene | ORF (AA) | Full Length | Signal Peptide (AA) | pI/Mw (kDa) | NPS | NCBI Blast Hit to Anoplophora glabripennis (Reference/Name) | E Value | Identity (%) |
|---|---|---|---|---|---|---|---|---|
| UGT1 | 517 | Yes | 20 | 8.88/58.57 | 129|174|239|509 | XP_018579880.1 UDP-glucuronosyltransferase 2B10 | 0.0 | 83 |
| UGT2 | 517 | Yes | 18 | 8.93/58.86 | 107|415|416|452 | XP_023312103.1 UDP-glucuronosyltransferase 2B15 | 0.0 | 79 |
| UGT3 | 517 | Yes | 18 | 8.79/59.32 | 65|121|220|397 | XP_018561507.1 UDP-glucuronosyltransferase 2B7 | 0.0 | 68 |
| UGT4 | 520 | Yes | 18 | 7.05/59.53 | 64|233 | XP_018563298.1 UDP-glucuronosyltransferase 2B10 | 0.0 | 87 |
| UGT5 | 512 | Yes | 18 | 8.70/58.16 | 120|230|456 | XP_018563256.1 UDP-glucuronosyltransferase 2B7-like | 0.0 | 84 |
| UGT6 | 516 | Yes | 22 | 8.96/58.92 | 119|175|240 | XP_018579878.1 UDP-glucuronosyltransferase 2B31 | 0.0 | 83 |
| UGT7 | 519 | Yes | 19 | 9.24/59.03 | 66|81|88|222|232|419 | XP_018561504.1 UDP-glucuronosyltransferase 2B37 isoform X1 | 0.0 | 56 |
| UGT8 | 526 | Yes | 27 | 9.14/61.25 | 91|246|429|517 | XP_018561622.1 UDP-glucuronosyltransferase 2B31 | 0.0 | 85 |
| UGT9 | 512 | Yes | 17 | 8.82/58.39 | 233|278|323 | XP_018570348.1 UDP-glucuronosyltransferase 1-8 | 0.0 | 81 |
| UGT10 | 523 | Yes | 19 | 8.73/58.99 | 50|94|128|173|238|273 | XP_018579876.1 UDP-glucuronosyltransferase 2B15 | 0.0 | 76 |
| UGT11 | 514 | Yes | 17 | 9.21/58.27 | 63|235 | XP_018579879.1 UDP-glucuronosyltransferase 1-8 | 0.0 | 84 |
| UGT12 | 523 | Yes | 18 | 8.29/60.04 | 226|517 | XP_018564526.1 UDP-glucuronosyltransferase 2B13-like | 0.0 | 91 |
| UGT13 | 533 | Yes | 22 | 9.09/61.38 | 69|177|243|274|334|419 | XP_018568770.1 UDP-glucuronosyltransferase 2B19 isoform X1 | 0.0 | 79 |
| UGT14 | 499 | Yes | 20 | 7.36/56.98 | 66|169|234|302|417|461 | XP_018565808.1 UDP-glucuronosyltransferase 2B16-like isoform X1 | 0.0 | 70 |
| UGT15 | 518 | Yes | 18 | 8.96/59.06 | 65 | XP_018573571.1 UDP-glucuronosyltransferase 2B7-like | 0.0 | 71 |
| UGT16 | 515 | Yes | 20 | 9.28/59.34 | 170|235|409 | XP_018563264.1 UDP-glucuronosyltransferase 2B37 | 0.0 | 73 |
| UGT17 | 516 | Yes | 17 | 8.91/58.43 | 64|120|219 | XP_018561507.1 UDP-glucuronosyltransferase 2B7 | 0.0 | 66 |
| UGT18 | 517 | Yes | 18 | 8.95/58.41 | 49|62|65|72|121 | XP_018561507.1 UDP-glucuronosyltransferase 2B7 | 0.0 | 67 |
| UGT19 | 522 | Yes | 17 | 9.19/59.78 | 64|225 | XP_018573569.1 UDP-glucuronosyltransferase 2B7 | 0.0 | 95 |
| UGT20 | 527 | Yes | 19 | 9.41/59.13 | 101|171|188|197|236 | XP_018562714.1 UDP-glucuronosyltransferase | 0.0 | 73 |
| UGT21 | 516 | Yes | 18 | 9.14/58.71 | 109|127|172|189|237 | XP_018579881.1 UDP-glucuronosyltransferase 2B7-like | 0.0 | 81 |
| UGT22 | 514 | Yes | 17 | 8.95/59.13 | 48|119|229|289|455 | XP_018562715.1 UDP-glucuronosyltransferase 2A3 isoform X1 | 0.0 | 84 |
| UGT23 | 522 | Yes | 20 | 6.44/60.65 | 64|85|173|285|336|424 | XP_018566903.1 UDP-glucuronosyltransferase 2B15-like | 0.0 | 79 |
| UGT24 | 527 | Yes | 21 | 8.65/59.92 | 52|82|239 | XP_018561400.1 UDP-glucuronosyltransferase 2C1-like | 0.0 | 96 |
| UGT25 | 517 | Yes | 18 | 9.01/59.23 | 65|122|236 | XP_018572801.1 UDP-glucuronosyltransferase 2B7 | 0.0 | 91 |
| UGT26 | 516 | Yes | 19 | 8.42/59.11 | 50|457 | XP_018570251.1 UDP-glucuronosyltransferase 1-6-like | 0.0 | 74 |
| UGT27 | 523 | Yes | 29 | 9.15/59.91 | 2|12|131|241|508 | XP_018563266.1 UDP-glucuronosyltransferase 2B1 | 0.0 | 79 |
| UGT28 | 511 | Yes | 17 | 6.58/58.41 | 64|166 | XP_018561499.1 UDP-glucuronosyltransferase 2B7-like | 0.0 | 70 |
| UGT29 | 521 | Yes | 19 | 6.14/59.21 | 65|123|238|245|465 | XP_018561622.1 UDP-glucuronosyltransferase 2B31 | 2 × 10−163 | 46 |
| UGT30 | 521 | Yes | 20 | 6.35/59.69 | 127|408 | XP_018561584.1 uncharacterized protein LOC108903775 | 0.0 | 78 |
| UGT31 | 520 | Yes | 20 | 7.63/58.90 | 51 | XP_018569262.1 UDP-glucuronosyltransferase 2B7 | 0.0 | 77 |
| UGT32 | 515 | Yes | 20 | 9.44/58.56 | 14|106|236|281|326 | XP_018570347.1 UDP-glucuronosyltransferase 1-1 isoform X2 | 0.0 | 87 |
| UGT33 | 521 | Yes | 24 | 9.24/58.90 | 112|176|242|287|332 | XP_018570346.1 UDP-glucuronosyltransferase 1-3 isoform X1 | 0.0 | 87 |
| UGT34 | 519 | Yes | 20 | 9.03/59.13 | 66|73|148|169|211 | XP_023310147.1 UDP-glucuronosyltransferase 2B16-like isoform X1 | 0.0 | 80 |
| UGT35 | 524 | No | XP_018579876.1 UDP-glucuronosyltransferase 2B15 | 0.0 | 67 | |||
| UGT36 | 522 | No | XP_018561504.1 UDP-glucuronosyltransferase 2B37 isoform X1 | 0.0 | 77 | |||
| UGT37 | 466 | No | XP_018565808.1 UDP-glucuronosyltransferase 2B16-like isoform X1 | 0.0 | 75 | |||
| UGT38 | 341 | No | XP_018563264.1 UDP-glucuronosyltransferase 2B37 | 2 × 10−174 | 70 | |||
| UGT39 | 338 | No | XP_018561507.1 UDP-glucuronosyltransferase 2B7 | 3 × 10−150 | 65 | |||
| UGT40 | 340 | No | XP_018568773.2 UDP-glucuronosyltransferase 2B9 | 3 × 10−175 | 73 | |||
| UGT41 | 292 | No | XP_018561504.1 UDP-glucuronosyltransferase 2B37 isoform X1 | 5 × 10−152 | 73 | |||
| UGT42 | 230 | No | XP_018563273.1 UDP-glucuronosyltransferase 2B9-like | 3 × 10−144 | 86 | |||
| UGT43 | 250 | No | XP_018579876.1 UDP-glucuronosyltransferase 2B15 | 9 × 10−128 | 76 | |||
| UGT44 | 448 | No | XP_018563300.1 UDP-glucuronosyltransferase 2B33-like | 0.0 | 81 | |||
| UGT45 | 275 | No | XP_018579881.1 UDP-glucuronosyltransferase 2B7-like | 1 × 10−153 | 76 | |||
| UGT46 | 413 | No | XP_018563264.1 UDP-glucuronosyltransferase 2B37 | 0.0 | 77 | |||
| UGT47 | 228 | No | XP_018561584.1 uncharacterized protein LOC108903775 | 2 × 10−121 | 78 | |||
| UGT48 | 428 | No | XP_023310231.1 UDP-glucuronosyltransferase 2B33-like | 0.0 | 71 | |||
| UGT49 | 453 | No | XP_018579876.1 UDP-glucuronosyltransferase 2B15 | 0.0 | 82 | |||
| UGT50 | 436 | No | XP_018579876.1 UDP-glucuronosyltransferase 2B15 | 0.0 | 79 | |||
| UGT51 | 350 | No | XP_018561520.1 UDP-glucuronosyltransferase 2B7-like | 0.0 | 83 | |||
| UGT52 | 287 | No | XP_018570348.1 UDP-glucuronosyltransferase 1-8 | 9 × 10−139 | 72 | |||
| UGT53 | 316 | No | XP_018563264.1 UDP-glucuronosyltransferase 2B37 | 0.0 | 89 | |||
| UGT54 | 276 | No | XP_018561499.1 UDP-glucuronosyltransferase 2B7-like | 1 × 10−84 | 49 | |||
| UGT55 | 221 | No | XP_018568773.2 UDP-glucuronosyltransferase 2B9 | 5 × 10−111 | 73 | |||
| UGT56 | 169 | No | XP_018561622.1 UDP-glucuronosyltransferase 2B31 | 6 × 10−30 | 39 | |||
| UGT57 | 177 | No | XP_018570251.1 UDP-glucuronosyltransferase 1-6-like | 7 × 10−77 | 65 | |||
| UGT58 | 164 | No | XP_018561504.1 UDP-glucuronosyltransferase 2B37 isoform X1 | 3 × 10−45 | 53 | |||
| UGT59 | 136 | No | XP_018561622.1 UDP-glucuronosyltransferase 2B31 | 8 × 10−45 | 52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, N.; Wang, Z.; Xiao, H.; Lu, T.; Liu, N. Identification and Characterization of UDP-Glycosyltransferase Genes in a Cerambycid Beetle, Pharsalia antennata Gahan, 1894 (Coleoptera: Cerambycidae). Diversity 2022, 14, 348. https://doi.org/10.3390/d14050348
Yin N, Wang Z, Xiao H, Lu T, Liu N. Identification and Characterization of UDP-Glycosyltransferase Genes in a Cerambycid Beetle, Pharsalia antennata Gahan, 1894 (Coleoptera: Cerambycidae). Diversity. 2022; 14(5):348. https://doi.org/10.3390/d14050348
Chicago/Turabian StyleYin, Ningna, Zhengquan Wang, Haiyan Xiao, Tingting Lu, and Naiyong Liu. 2022. "Identification and Characterization of UDP-Glycosyltransferase Genes in a Cerambycid Beetle, Pharsalia antennata Gahan, 1894 (Coleoptera: Cerambycidae)" Diversity 14, no. 5: 348. https://doi.org/10.3390/d14050348
APA StyleYin, N., Wang, Z., Xiao, H., Lu, T., & Liu, N. (2022). Identification and Characterization of UDP-Glycosyltransferase Genes in a Cerambycid Beetle, Pharsalia antennata Gahan, 1894 (Coleoptera: Cerambycidae). Diversity, 14(5), 348. https://doi.org/10.3390/d14050348