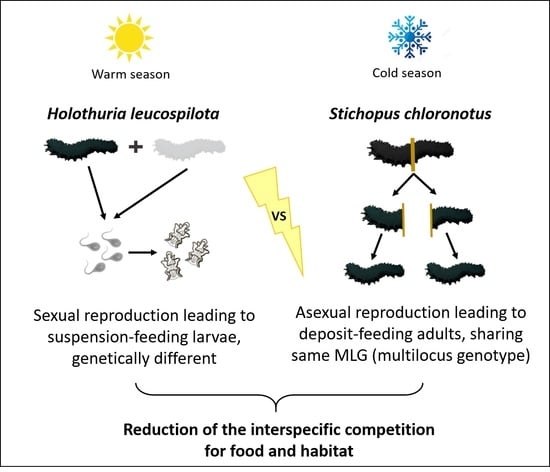
Sex or Fission? Genetics Highlight Differences in Reproductive Strategies of Two Sympatric Fissiparous Sea Cucumber Species in Reunion Island (Southwestern Indian Ocean)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Design
2.1.1. Spatial Sampling

2.1.2. Temporal Sampling of Both Species
2.2. Laboratory Steps
2.3. Data Analyses
2.3.1. Clonal Identification and Propagation
2.3.2. Genetic Diversity
2.3.3. Population Structure and Differentiation
3. Results
3.1. MLG and Clone Identification
3.1.1. Clonal Diversity of Holothuria leucospilota
3.1.2. Clonal Diversity of Stichopus chloronotus
3.2. Clonal Propagation of Stichopus chloronotus through Space and Time
3.3. Population Structure and Differentiation
4. Discussion
4.1. Importance of the Sexual Reproduction for Holothuria leucospilota
4.2. Importance of Asexual Reproduction for Stichopus chloronotus
4.3. Differences in Reproductive Strategies in Two Sympatric Sea Cucumber Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beliaev, G.M. Deep Sea Ocean Trenches and Their Fauna; Nauka: Moscow, Russia, 1989; ISBN 978-5-02-005276-5. [Google Scholar]
- Kuhnz, L.A.; Ruhl, H.A.; Huffard, C.L.; Smith, K.L. Rapid Changes and Long-Term Cycles in the Benthic Megafaunal Community Observed over 24 years in the Abyssal Northeast Pacific. Prog. Oceanogr. 2014, 124, 1–11. [Google Scholar] [CrossRef]
- Wolfe, K.; Davey, M. Localised High-Density Population of a Sea Cucumber on a Malaysian Coral Reef. Coral Reefs 2020, 39, 33–38. [Google Scholar] [CrossRef]
- Conde, E.C.; Diaz, H.; Sambrani, A. Disintegration of Holothurian Faecal Pellets in Beds of the Seagrass Thalassia testudinum. J. Coast. Res. 1991, 7, 853–862. [Google Scholar]
- WoRMS Editorial Board. World Register of Marine Species. 2021. Available online: https://www.marinespecies.org (accessed on 20 October 2021).
- Purcell, S.S.; Conand, C.; Uthicke, S.; Byrne, M. Ecological Roles of Exploited Sea Cucumbers. Oceanogr. Mar. Biol. Annu. Rev. 2016, 54, 367–386. [Google Scholar] [CrossRef]
- Toral-Granda, V.; Lovatelli, A.; Vasconcellos, M. Sea Cucumbers: A Global Review of Fisheries and Trade; FAO Fisheries and Aquaculture Technical Paper; FAO: Rome, Italy, 2008; ISBN 978-92-5-106079-7. [Google Scholar]
- Conand, C. The Fishery Resources of Pacific Island Countries: Holothurians; FAO Fisheries and Wquaculture Technical Paper; FAO: Rome, Italy, 1990; Volume 272, 143p, Available online: https://scholar.google.fr/scholar?cluster=8908028655790245485&hl=fr&as_sdt=0,5 (accessed on 15 March 2023).
- Conand, C. Tropical Sea Cucumber Fisheries: Changes during the Last Decade. Mar. Pollut. Bull. 2018, 133, 590–594. [Google Scholar] [CrossRef]
- Hamel, J.F.; Eeckhaut, I.; Conand, C.; Sun, J.; Caulier, G.; Mercier, A. Global Knowledge on the Commercial Sea Cucumber Holothuria scabra. Adv. Mar. Biol. 2022, 91, 1–286. [Google Scholar]
- Yang, H.; Hamel, J.F.; Mercier, A. The Sea Cucumber Apostichopus Japonicus: History, Biology and Aquaculture; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Rahman, M.A.; Yusoff, F. Sea Cucumber Fisheries: Market Potential, Trade, Utilization and Challenges for Expanding the Production in the South-East Asia. Int. J. Adv. Chem. Eng. Biol. Sci. 2017, 4, 26–30. [Google Scholar] [CrossRef]
- Friedman, K.; Eriksson, H.; Tardy, E.; Pakoa, K. Management of Sea Cucumber Stocks: Patterns of Vulnerability and Recovery of Sea Cucumber Stocks Impacted by Fishing. Fish Fish. 2011, 12, 75–93. [Google Scholar] [CrossRef]
- Rahman, M.A.; Yusoff, F.; Arshad, A. Sea Cucumber Fisheries: Global Status, Culture, Management and Extinction Risks. Int. J. Chem. Environ. Biol. Sci. 2015, 3, 344–348. [Google Scholar]
- Purcell, S.W.; Samyn, Y.; Conand, C. Commercially Important Sea Cucumbers of the World. In FAO Species Catalogue for Fishery Purposes; FAO: Rome, Italy, 2012; ISBN 978-92-5-106719-2. [Google Scholar]
- Purcell, S.W. Value, Market Preferences and Trade of Beche-De-Mer from Pacific Island Sea Cucumbers. PLoS ONE 2014, 9, e95075. [Google Scholar] [CrossRef]
- Drumm, D.J.; Loneragan, N.R. Reproductive Biology of Holothuria leucospilota in the Cook Islands and the Implications of Traditional Fishing of Gonads on the Population. N. Z. J. Mar. Freshw. Res. 2005, 39, 141–156. [Google Scholar] [CrossRef]
- Adibpour, N.; Nasr, F.; Nematpour, F.; Shakouri, A.; Ameri, A. Antibacterial and Antifungal Activity of Holothuria leucospilota Isolated From Persian Gulf and Oman Sea. Jundishapur J. Microbiol. 2014, 7, e8708. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, C.; Zheng, Q.; Wu, J.; Zhu, K.; Shen, X.; Cao, J. Effect of Simulated Gastrointestinal Digestion in Vitro on the Antioxidant Activity, Molecular Weight and Microstructure of Polysaccharides from a Tropical Sea Cucumber (Holothuria leucospilota). Food Hydrocoll. 2019, 89, 735–741. [Google Scholar] [CrossRef]
- Gozari, M.; Bahador, N.; Jassbi, A.R.; Mortazavi, M.S.; Eftekhar, E. Antioxidant and Cytotoxic Activities of Metabolites Produced by a New Marine Streptomyces sp. Isolated from the Sea Cucumber Holothuria leucospilota. Iran. J. Fish Sci. 2018, 17, 413–426. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, Y.; Xu, B.; Wu, J.; Zhang, L.; Gao, M.; Zheng, S.; Wang, A.; Zhang, C.; Chen, L.; et al. Acidic Mucopolysaccharide from Holothuria leucospilota Has Antitumor Effect by Inhibiting Angiogenesis and Tumor Cell Invasion In Vivo and In Vitro. Cancer Biol. Ther. 2009, 8, 1489–1499. [Google Scholar] [CrossRef]
- Conand, C.; Trentin, F.; Mulochau, T. Marine Biodiversity of La Reunion Island. West. Ind. Ocean J. Mar. Sci. 2018, 17, 111–127. [Google Scholar]
- Conand, C.; Morel, C.; Mussard, R. A New Study Asexual Reproduction in Holothurians: Fission in Holothuria leucospilota Populations on Reunion Island in the Indian Ocean. SPC Beche. Mer. Inf. Bull. 1997, 9, 5–11. [Google Scholar]
- Conand, C.; Armand, J.; Dijoux, N.; Garryer, J. Fission in a Population of Stichopus chloronotus on Reunion Island, Indian Ocean. SPC Beche. Mer. Inf. Bull. 1998, 10, 15–24. [Google Scholar]
- Conand, C.; Mangion, P. Sea Cucumbers on La Reunion Island Fringing Reefs: Diversity, Distribution, Abundance and Structure of the Populations. SPC Beche. Mer. Inf. Bull. 2002, 17, 27–34. [Google Scholar]
- Cuvillier, A. Dynamique et Fonctionnement Des Herbiers Marins Dans Un Complexe Récifal Anthropisé (Île de La Réunion, Océan Indien). Ph.D. Thesis, Université de La Réunion, La Réunion, France, 2016. [Google Scholar]
- Dolmatov, I.Y. Asexual Reproduction in Holothurians. Sci. World J. 2014, 2014, 527234. [Google Scholar] [CrossRef]
- Purwati, P. Fissiparity in Holothuria leucospilota from Tropical Darwin Waters, Northern Australia. SPC Beche. Mer. Inf. Bull. 2004, 20, 26–33. [Google Scholar]
- Dai, G.; Li, Z.B.; Shangguan, J.B.; Ning, Y.F.; Deng, H.W.; Yuan, Y.; Huang, Y.S.; Yang, H.; Lu, J. Development and Characterization of Polymorphic Microsatellite Loci in the Sea Cucumber Holothuria leucospilota. Genet. Mol. Res. 2015, 14, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, J.B.; Li, Z.B.; Ning, Y.F.; Huang, Y.S.; Yuan, Y.; Lu, J.; Li, B.B.; Mao, X.Q. Screening and Characterization of Novel Polymorphic Microsatellite Markers from Sea Cucumber Holothuria leucospilota. Genet. Mol. Res. 2015, 14, 6555–6560. [Google Scholar] [CrossRef] [PubMed]
- Pirog, A.; Gélin, P.; Bédier, A.; Bianchetti, G.; Georget, S.; Frouin, P.; Magalon, H. Clonal Structure through Space and Time: High Stability in the Holothurian Stichopus chloronotus (Echinodermata). Ecol. Evol. 2017, 7, 7534–7547. [Google Scholar] [CrossRef] [PubMed]
- Pierrat, J.; Magalon, H.; Libaud, N.; Oury, N. Isolation and Characterization of 21 Microsatellite Loci for the Sea Cucumber Holothuria (Halodeima) atra (Echinodermata, Holothuroidea) Reveal Low Asexual Propagation through Time in Reunion Island (Southwestern Indian Ocean). Mol. Biol. Rep. 2022, 50, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Pierrat, J.; Libaud, N.; Magalon, H.; Oury, N. Isolation and Characterization of 24 Microsatellite Loci from One of the Most Widespread Sea Cucumber Holothuria (Mertensiothuria) leucospilota (Echinodermata, Holothuroidea). Conserv. Genet. Resour. 2022, 14, 389–390. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.r-project.org/ (accessed on 15 October 2022).
- Dorken, M.E.; Eckert, C.G. Severely Reduced Sexual Reproduction in Northern Populations of a Clonal Plant, Decodon Verticillatus (Lythraceae): Reduced Sexuality in Northern Decodon. J. Ecol. 2001, 89, 339–350. [Google Scholar] [CrossRef]
- Wright, S. Evolution in Mendelian Populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices Version 2.9.3.2, Updated from Goudet 1995. 2001. Available online: https://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 15 March 2023).
- Raymond, M.; Rousset, F. GenePop: Population Genetics Software for Exact Tests and Ecumenism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A Complete Re-Implementation of the Genepop Software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant Analysis of Principal Components: A New Method for the Analysis of Genetically Structured Populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Conand, C. Asexual Reproduction by Fission in Holothuria atra Variability of Some Parameters in Populations from the Tropical Lndo-Pacific. Oceanol. Acta 1995, 19, 209–216. [Google Scholar]
- Conand, C. Monitoring a Fissiparous Population of Holothuria atra on a Fringing Reef on Reunion Island (Indian Ocean). SPC Beche. Mer. Inf. Bull. 2004, 20, 22–26. [Google Scholar]
- Gianasi, B.L.; Verkaik, K.; Hamel, J.-F.; Mercier, A. Novel Use of PIT Tags in Sea Cucumbers: Promising Results with the Commercial Species Cucumaria frondosa. PLoS ONE 2015, 10, e0127884. [Google Scholar] [CrossRef]
- Gaudron, S.M.; Kohler, S.A.; Conand, C. Reproduction of the Sea Cucumber Holothuria leucospilota in the Western Indian Ocean: Biological and Ecological Aspects. Invertebr. Reprod. Dev. 2008, 51, 19–31. [Google Scholar] [CrossRef]
- Ong Che, R.G. Reproductive Cycle of Holothuria leucospilota Brandt (Echinodermata: Holothuridea) in Hong Kong and the Role of Body Tissues in Reproduction. Asian Mar. Biol. 1990, 7, 115–132. [Google Scholar]
- Franklin, S.E. The Reproductive Biology and Some Aspects of the Population Ecology of the Holothurians Holothuria leucospilota and Stichopus chloronotus. Ph.D. Thesis, University of Sydney, Sydney, Australia, 1980. [Google Scholar]
- Purwati, P.; Luong-van, J.T. Sexual Reproduction in a Fissiparous Holothurian Species, Holothuria leucospilota Clark 1920 (Echinodermata: Holothuroidea). SPC Beche. Mer. Inf. Bull. 2003, 18, 33–38. [Google Scholar]
- Uthicke, S.; Benzie, J.A.H.; Ballment, E. Population Genetics of the Fissiparous Holothurian Stichopus chloronotus (Aspidochirotida) on the Great Barrier Reef, Australia. Coral Reefs 1999, 18, 123–132. [Google Scholar] [CrossRef]
- Uthicke, S.; Conand, C. Amplified Fragment Length Polymorphism (AFLP) Analysis Indicates the Importance of Both Asexual and Sexual Reproduction in the Fissiparous Holothurian Stichopus chloronotus (Aspidochirotida) in the Indian and Pacific Ocean. Coral Reefs 2005, 24, 103–111. [Google Scholar] [CrossRef]
- Gélin, P.; Fauvelot, C.; Mehn, V.; Bureau, S.; Rouzé, H.; Magalon, H. Superclone Expansion, Long-Distance Clonal Dispersal and Local Genetic Structuring in the Coral Pocillopora damicornis Type β in Reunion Island, South Western Indian Ocean. PLoS ONE 2017, 12, e0169692. [Google Scholar] [CrossRef] [PubMed]
- Uthicke, S. Seasonality of Asexual Reproduction in Holothuria (Halodeima) atra, H. (H.) edulis and Stichopus chloronotus (Holothuroidea: Aspidochirotida) on the Great Barrier Reef. Mar. Biol. 1997, 129, 435–441. [Google Scholar] [CrossRef]
- Bonham, K.; Held, E.E. Ecological Observations on the Sea Cucumbers Holothuria atra and H. leucospilota at Rongelap Atoll, Marshall Islands. Pac. Sci. 1963, 17, 305–314. [Google Scholar]
- Menge, B.A. Brood or Broadcast? The Adaptive Significance of Different Reproductive Strategies in the Two Intertidal Sea Stars Leptasterias hexactis and Pisaster ochraceus. Mar. Biol. 1975, 31, 87–100. [Google Scholar] [CrossRef]
- Schmidt-Roach, S.; Miller, K.J.; Woolsey, E.; Gerlach, G.; Baird, A.H. Broadcast Spawning by Pocillopora Species on the Great Barrier Reef. PLoS ONE 2012, 7, e50847. [Google Scholar] [CrossRef]
- Keunecke, K.A.; D’Incao, F.; Verani, J.R.; Vianna, M. Reproductive Strategies of Two Sympatric Swimming Crabs Callinectes danae and Callinectes ornatus (Crustacea: Portunidae) in an Estuarine System, South-Eastern Brazil. J. Mar. Biol. Assoc. UK 2012, 92, 343–347. [Google Scholar] [CrossRef]
- de Caralt, S.; González, J.; Turon, X.; Uriz, M.J. Reproductive Strategies of Two Common Sympatric Mediterranean Sponges: Dysidea avara (Dictyoceratida) and Phorbas tenacior (Poecilosclerida). PeerJ 2018, 6, e5458. [Google Scholar] [CrossRef] [PubMed]
- Mercier, A.; Battaglene, S.C.; Hamel, J.-F. Settlement Preferences and Early Migration of the Tropical Sea Cucumber Holothuria scabra. J. Exp. Mar. Biol. Ecol. 2000, 249, 89–110. [Google Scholar] [CrossRef] [PubMed]



| Panel | Locus | Primer Sequence (5′–3′) | Dye | Specific Size Range (bp) |
|---|---|---|---|---|
| 1 | Hl21 | F: TGTTTCACGAATGAATGAACG | 6-FAM | 220–320 |
| R: GCTTGTAAAGCCATTTGTACCTT | ||||
| Hl04 | F: CCCAGAAGCTCTGGAACATT | VIC | 170–184 | |
| R: TGCTATGTAAACTGAAGCCAAA | ||||
| Hl10 | F: AAACGTCCTCGATTGACAGC | NED | 137–165 | |
| R: TCTGCTAGCCAAATTACAGGG | ||||
| Hl19 | F: GCCGATTCCTTTGAACATTA | 6-FAM | 91–132 | |
| R: AATTGGTTGGAAACTGGGAC | ||||
| 2 | Hl23 | F: GGTCAAAGAACCTGCAGACA | 6-FAM | 238–274 |
| R: CCCGACTCAAGCATTACTTAAA | ||||
| Hl06 | F: CGTCACGTTACGAATGGTACTC | VIC | 192–208 | |
| R: TTGGCGCATTTCCTTACAAT | ||||
| Hl15 | F: TCCAAGTATGAGATCCGTCG | NED | 144–168 | |
| R: CAGTCCTTGCCGAATGCT | ||||
| Hl08 | F: AATCTGGTCTGCTTTCAGGA | 6-FAM | 126–138 | |
| R: AAACTGCCTGGGTAAGTCTGT | ||||
| 3 | Hl01 | F: ATCGTGTTTACAAGCTAGGCG | 6-FAM | 239–291 |
| R: AGATGTTGCTAGACCACTGCAT | ||||
| Hl05 | F: ATTGGCAGGCAAGGAATCTA | VIC | 166–180 | |
| R: GTCTATGTCGCCTGATGGCT | ||||
| Hl03 | F: TTTCATTATGTTGCACCCACC | NED | 134–156 | |
| R: TGTAAAGCACAACTTTGCGTG | ||||
| Hl14 | F: TGCAGTGCCATATCCAACAT | 6-FAM | 129–149 | |
| R: TTCTTTCATCCTCTCGGCAT | ||||
| 4 | Hl12 | F: CAGCACATAGTATACTGCATTCCC | 6-FAM | 268–278 |
| R: AAATTCCGTCACTGCAAAGAA | ||||
| Hl16 | F: TAGAAATCCTTTCCGCGTGT | VIC | 200–228 | |
| R: GATGCCCTCGGATTGTATGT | ||||
| Hl13 | F: CAAGTGTTCCAAACTGGGCT | NED | 133–165 | |
| R: TCTTCGGGAAGTGTTAGTTGC | ||||
| Hl20 | F: CGGGTGCAGAAAGTACCCTA | 6-FAM | 130–174 | |
| R: GGTTCCAACTCCCTGGTCTT | ||||
| 5 | Hl24 | F: GTTAATACGTCAAGTAACGTAGACTGC | 6-FAM | 294–304 |
| R: TTCCTTCTTATTTGGCGAGC | ||||
| Hl11 | F: GAACTAACAGCCACGATTGG | VIC | 201–215 | |
| R: CGCATAAACTGTGAAGAAGATCC | ||||
| Hl22 | F: TCAGGTGATTAGTAGCTCAGCAAG | 6-FAM | 143–185 | |
| R: CCAACTTTGAGAAGGAACGG | ||||
| Hl02 | F: CCGTAAGGCATCGAGTGTG | NED | 130–134 | |
| R: ACATTCGAGAAGGAAGCTTGA | ||||
| 6 | Hl17 | F: GAATCTTATAATCCCTTGGTTCTCA | 6-FAM | 273–321 |
| R: TCGATCTAACATATAGAATCGTTGG | ||||
| Hl07 | F: AACTGGCTTCAATGACACTACG | VIC | 205–221 | |
| R: TTGATCGCTTGGTTATTGAGTT | ||||
| Hl09 | F: GAATAATCACAAGTTTGACGGC | NED | 145–189 | |
| R: TAATCTTGAGAAGCCGGTGT | ||||
| Hl18 | F: CACGAACAGATTTCTTTGTTGTTC | 6-FAM | 132–174 | |
| R: TGTGGAAGATCACGGGTAAG |
| Panel | Locus | Primer Sequence (5′–3′) | Dye | Specific Size Range (bp) |
|---|---|---|---|---|
| 1 | Sc10 | F: CGCCTCTAATCTCAAATTGTCG | 6-FAM | 142–164 |
| R: TGCGGTCTTCCTTGTCTC | ||||
| Sc09 | F: CCAATGCTTTGATTCCAGG | VIC | 200–206 | |
| R: CCAACTTGCACATATTGAG | ||||
| Sc43 | F: CGTGACATACAACTTCCTAGC | 6-FAM | 233–239 | |
| R: GAGATCACTTAGAGTTACGC | ||||
| Sc01 | F: CGGGAAGCATTAAAAGTCGC | VIC | 323–326 | |
| R: GCGATACGGATCCTTGTGG | ||||
| 2 | Sc24 | F: CGTGGTTAAATTCCTAGGTATAGAG | 6-FAM | 148–158 |
| R: CTGGAATAAACCTGATGTAC | ||||
| Sm007 | F: CACCGCTTTGAATTTGTAG | VIC | 172–176 | |
| R: ACTGTAGGCAATGAATGA | ||||
| Sc29 | F: GTAGCCCATAAATCATTG | NED | 212–218 | |
| R: GACCAACCCACACAGCAAG | ||||
| Sc33 | F: CTGGTTCGGATTCACATAG | 6-FAM | 260–266 | |
| R: CTACTTACGGTGAAACTTCC | ||||
| Sm014 | F: CACGGACAGTGGTCACAAG | VIC | 355–365 | |
| R: TGAGATAGAGCGTTTACGAG |
| Season | Site | %NA | N | NMLG | R | Na | Np | Ho | He | FIS |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | MNS | 58.33 | 10 | 10 | 1 | 6.17 ± 0.58 | 0.96 ± 0.24 | 0.39 ± 0.05 | 0.82 ± 0.03 | 0.53 *** ± 0.05 |
| CAP | 50.00 | 6 | 6 | 1 | 4.96 ± 0.39 | 0.50 ± 0.17 | 0.41 ± 0.06 | 0.82 ± 0.03 | 0.48 *** ± 0.07 | |
| PLA | 37.50 | 15 | 15 | 1 | 7.04 ± 0.57 | 1.08 ± 0.21 | 0.41 ± 0.04 | 0.80 ± 0.03 | 0.50 *** ± 0.04 | |
| PTE | 41.67 | 7 | 7 | 1 | 5.58 ± 0.49 | 0.46 ± 0.16 | 0.45 ± 0.06 | 0.80 ± 0.04 | 0.43 *** ± 0.07 | |
| TE | 33.33 | 16 | 16 | 1 | 7.38 ± 0.65 | 0.92 ± 0.18 | 0.33 ± 0.04 | 0.80 ± 0.03 | 0.58 *** ± 0.04 | |
| S2 | MNS | 4.17 | 23 | 23 | 1 | 10.38 ± 0.80 | 0.96 ± 0.20 | 0.57 ± 0.04 | 0.82 ± 0.02 | 0.31 *** ± 0.04 |
| CAP | 0.00 | 12 | 12 | 1 | 7.67 ± 0.64 | 0.42 ± 0.18 | 0.54 ± 0.05 | 0.79 ± 0.03 | 0.31 *** ± 0.05 | |
| PLA | 0.00 | 24 | 24 | 1 | 10.46 ± 0.82 | 0.75 ± 0.24 | 0.52 ± 0.04 | 0.82 ± 0.02 | 0.37 *** ± 0.04 | |
| PTE | 0.00 | 12 | 12 | 1 | 8.08 ± 0.54 | 0.50 ± 0.16 | 0.55 ± 0.05 | 0.83 ± 0.02 | 0.34 *** ± 0.05 | |
| TE | 4.17 | 23 | 23 | 1 | 10.17 ± 0.87 | 0.63 ± 0.19 | 0.53 ± 0.04 | 0.80 ± 0.003 | 0.34 *** ± 0.04 | |
| S3 | MNS | 0.00 | 24 | 24 | 1 | 10.46 ± 0.70 | 1.13 ± 0.23 | 0.53 ± 0.05 | 0.83 ± 0.02 | 0.37 *** ± 0.05 |
| CAP | 0.00 | 12 | 12 | 1 | 7.67 ± 0.49 | 0.33 ± 0.10 | 0.50 ± 0.05 | 0.82 ± 0.02 | 0.38 *** ± 0.06 | |
| PLA | 12.50 | 21 | 21 | 1 | 9.71 ± 0.62 | 0.58 ± 0.15 | 0.59 ± 0.04 | 0.81 ± 0.02 | 0.27 *** ± 0.04 | |
| PTE | 0.00 | 12 | 12 | 1 | 7.58 ± 0.58 | 0.29 ± 0.11 | 0.52 ± 0.05 | 0.81 ± 0.03 | 0.35 *** ± 0.05 | |
| TE | 0.00 | 24 | 24 | 1 | 10.54 ± 0.72 | 1.38 ± 0.26 | 0.53 ± 0.04 | 0.83 ± 0.03 | 0.36 *** ± 0.04 |
| Season | Site | %NA | N | NMLG | R | Na | Np | Ho | He | FIS |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | PAS | 79.17 | 5 | 4 | 0.75 | 1.78 ± 0.22 | 0.11 ± 0.11 | 0.40 ± 0.13 | 0.28 ± 0.08 | −0.44 *** ± 0.12 |
| TE | 66.67 | 8 | 3 | 0.29 | 1.78 ± 0.32 | 0.11 ± 0.11 | 0.17 ± 0.10 | 0.16 ± 0.06 | −0.02 NS ± 0.21 | |
| ES | 33.33 | 16 | 6 | 0.33 | 1.67 ± 0.17 | 0.22 ± 0.15 | 0.38 ± 0.13 | 0.26 ± 0.07 | −0.50 *** ± 0.19 | |
| S2 | PAS | 4.17 | 23 | 7 | 0.27 | 1.78 ± 0.22 | 0.11 ± 0.11 | 0.25 ± 0.09 | 0.23 ± 0.07 | −0.08 NS ± 0.12 |
| TE | 12.50 | 21 | 3 | 0.10 | 1.78 ± 0.22 | 0.11 ± 0.11 | 0.12 ± 0.10 | 0.10 ± 0.05 | −0.24 ** ± 0.24 | |
| ES | 0.00 | 24 | 2 | 0.04 | 1.56 ± 0.18 | 0.11 ± 0.11 | 0.41 ± 0.15 | 0.24 ± 0.08 | −0.73 *** ± 0.11 | |
| S3 | PAS | 8.33 | 22 | 6 | 0.24 | 2.00 ± 0.33 | 0.22 ± 0.15 | 0.26 ± 0.09 | 0.20 ± 0.06 | −0.29 *** ± 0.05 |
| TE | 0.00 | 24 | 3 | 0.09 | 1.78 ± 0.22 | 0.00 ± 0.00 | 0.13 ± 0.10 | 0.12 ± 0.05 | −0.02 NS ± 0.24 | |
| ES | 4.17 | 23 | 3 | 0.09 | 1.78 ± 0.15 | 0.11 ± 0.11 | 0.39 ± 0.12 | 0.26 ± 0.07 | −0.50 *** ± 0.10 |
| Season | S1cold | S2warm | S3cold | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | MNS | CAP | PLA | PTE | TE | MNS | CAP | PLA | PTE | TE | MNS | CAP | PLA | PTE | TE | |
| S1cold | MNS (10) | - | ||||||||||||||
| CAP (6) | 0.007 | - | ||||||||||||||
| PLA (15) | 0.015 | 0.007 | - | |||||||||||||
| PTE (7) | 0.033 | 0.028 | 0.017 | - | ||||||||||||
| TE (16) | 0.020 | 0.040 | 0.035 | 0.039 | - | |||||||||||
| S2warm | MNS (23) | 0.018 | 0.015 | 0.010 | 0.005 | 0.029 * | - | |||||||||
| CAP (12) | 0.045 | 0.036 | 0.048 * | 0.027 | 0.066 *** | 0.029 * | - | |||||||||
| PLA (24) | 0.020 | 0.017 | 0.015 | 0.002 | 0.024 | 0.014 | 0.019 | - | ||||||||
| PTE (12) | 0.033 | 0.021 | 0.026 | 0.006 | 0.045 * | 0.014 | 0.044 *** | 0.012 | - | |||||||
| TE (23) | 0.042 * | 0.028 | 0.032 * | 0.012 | 0.040 * | 0.012 | 0.038 *** | 0.013 | 0.009 | - | ||||||
| S3cold | MNS (24) | 0.029 | 0.031 | 0.028 * | 0.009 | 0.042 *** | 0.014 | 0.032 * | 0.016 | 0.007 | 0.013 | - | ||||
| CAP (12) | 0.027 | 0.025 | 0.023 | 0.008 | 0.042 | 0.006 | 0.016 | 0.010 | 0.021 | 0.013 | 0.007 | - | ||||
| PLA (21) | 0.026 | 0.015 | 0.012 | 0.009 | 0.045 *** | 0.009 | 0.015 | 0.012 | 0.021 | 0.017 | 0.013 | 0.005 | - | |||
| PTE (12) | 0.035 | 0.031 | 0.023 | 0.002 | 0.045 * | 0.015 | 0.023 | 0.009 | 0.019 | 0.011 | 0.004 | 0.003 | 0.011 | - | ||
| TE (24) | 0.018 | 0.032 | 0.017 | 0.009 | 0.029 * | 0.008 | 0.044 *** | 0.014 | 0.009 | 0.008 | 0.002 | 0.004 | 0.019 * | 0.008 | - | |
| Season | S1cold | S2warm | S3cold | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Site | PAS | TE | ES | PAS | TE | ES | PAS | TE | ES | |
| S1cold | PAS (5) | - | ||||||||
| TE (8) | 0.002 | - | ||||||||
| ES (16) | 0.081 ** | 0.113 *** | - | |||||||
| S2warm | PAS (23) | −0.002 | 0.064 | 0.195 *** | - | |||||
| TE (21) | 0.147 * | −0.016 | 0.220 *** | 0.138 *** | - | |||||
| ES (24) | 0.105 ** | 0.142 *** | −0.016 | 0.207 *** | 0.237 *** | - | ||||
| S3cold | PAS (22) | 0.017 | 0.068 * | 0.232 *** | −0.004 | 0.129 *** | 0.240 *** | - | ||
| TE (24) | 0.095 | −0.032 | 0.188 *** | 0.117 ** | −0.018 | 0.209 *** | 0.111 *** | - | ||
| ES (23) | 0.071 * | 0.115 *** | −0.020 | 0.189 *** | 0.219 *** | −0.007 | 0.231 *** | 0.188 *** | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierrat, J.; Oury, N.; Frouin, P.; Magalon, H. Sex or Fission? Genetics Highlight Differences in Reproductive Strategies of Two Sympatric Fissiparous Sea Cucumber Species in Reunion Island (Southwestern Indian Ocean). Diversity 2023, 15, 670. https://doi.org/10.3390/d15050670
Pierrat J, Oury N, Frouin P, Magalon H. Sex or Fission? Genetics Highlight Differences in Reproductive Strategies of Two Sympatric Fissiparous Sea Cucumber Species in Reunion Island (Southwestern Indian Ocean). Diversity. 2023; 15(5):670. https://doi.org/10.3390/d15050670
Chicago/Turabian StylePierrat, Joséphine, Nicolas Oury, Patrick Frouin, and Hélène Magalon. 2023. "Sex or Fission? Genetics Highlight Differences in Reproductive Strategies of Two Sympatric Fissiparous Sea Cucumber Species in Reunion Island (Southwestern Indian Ocean)" Diversity 15, no. 5: 670. https://doi.org/10.3390/d15050670
APA StylePierrat, J., Oury, N., Frouin, P., & Magalon, H. (2023). Sex or Fission? Genetics Highlight Differences in Reproductive Strategies of Two Sympatric Fissiparous Sea Cucumber Species in Reunion Island (Southwestern Indian Ocean). Diversity, 15(5), 670. https://doi.org/10.3390/d15050670