Disentangling the Anacondas: Revealing a New Green Species and Rethinking Yellows †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. DNA Isolation and Sequencing
2.3. Phylogenetic Analysis and Genetic Divergence
2.4. Divergence Time Estimation
2.5. Morphological Comparison of E. murinus between North and South
3. Results
3.1. Phylogenetics
3.1.1. Eunectes Overview
3.1.2. Yellow Anaconda Phylogenetics and Taxonomy
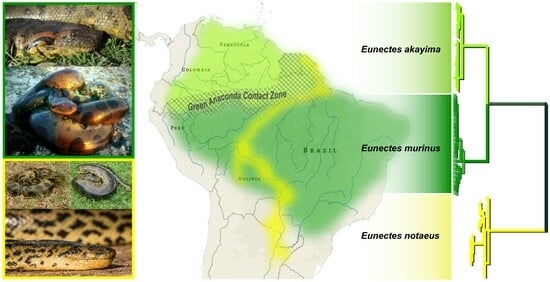
3.1.3. Green Anaconda Phylogenetics and Taxonomy
3.2. Divergence Time Estimation
4. Discussion
4.1. Taxonomic Implications for Yellow Anacondas

4.2. A New Species of Green Anaconda
Etymology
4.3. Paleogeographic Events Triggered the Origin of Large-Bodied Aquatic Snakes
4.4. Miocene Divergence of Northern and Southern Green Anacondas
4.5. The Arrival of the Pleistocene
4.6. Conservation Assessments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rull, V. Speciation Timing and Neotropical Biodiversity: The Tertiary-Quaternary Debate in the Light of Molecular Phylogenetic Evidence. Mol. Ecol. 2008, 17, 2722–2729. [Google Scholar] [CrossRef]
- Haffer, J. Hypotheses to Explain the Origin of Species in Amazonia. Braz. Biogeogr. 2008, 68, 917–947. [Google Scholar] [CrossRef]
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The Biodiversity of Species and Their Rates of Extinction, Distribution, and Protection. Science 2014, 344, 1246752. [Google Scholar] [CrossRef] [PubMed]
- Claramunt, S.; Cracraft, J. A New Time Tree Reveals Earth History’s Imprint on the Evolution of Modern Birds. Sci. Adv. 2015, 1, e1501005. [Google Scholar] [CrossRef]
- Rivas, J.A. Climate Changes and Speciation Pulses in a Nearly Flooded Continent: Tackling the Riddle of South America’s High Diversity. Ecotrópicos 2020, 32, 1–21. [Google Scholar] [CrossRef]
- Alroy, J. How Many Named Species Are Valid? Proc. Natl. Acad. Sci. USA 2002, 99, 3706–3711. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.; Schmink, M.; Abers, R.; Assad, E.; Humphreys Bebbington, D.; Eduaro, B.; Costa, F.; Durán Calisto, A.M.; Fearnside, P.M.; Garrett, R.; et al. The Amazon in Motion: Changing Politics, Development Strategies, Peoples, Landscapes, and Livehoods. In Amazon Assessment Report 2021, Part II; United Nations Sustainable Development Solutions Network: New York, NY, USA, 2021; pp. 14.1–14.65. [Google Scholar]
- Alencar, A.A.; Brando, P.M.; Asner, G.P.; Putz, F.E.; Alencar, A.A.; Brando, P.M.; Asner, G.P.; Putz, F.E. Landscape Fragmentation, Severe Drought, and the New Amazon Forest Fire Regime. Ecol. Appl. 2015, 25, 1493–1505. [Google Scholar] [CrossRef]
- Albert, J.S.; Carnaval, A.C.; Flantua, S.G.A.; Lohmann, L.G.; Ribas, C.C.; Riff, D.; Carrillo, J.D.; Fan, Y.; Figueiredo, J.J.P.; Guayasamin, J.M.; et al. Human Impacts Outpace Natural Processes in the Amazon. Science 2023, 379, eabo5003. [Google Scholar] [CrossRef]
- Velásquez Ramírez, M.G.; Barrantes, J.A.G.; Thomas, E.; Gamarra Miranda, L.A.; Pillaca, M.; Tello Peramas, L.D.; Bazán Tapia, L.R. Heavy Metals in Alluvial Gold Mine Spoils in the Peruvian Amazon. Catena 2020, 189, 104454. [Google Scholar] [CrossRef]
- Lages, A.S.; Miranda SA, F.; Ferreira SJ, F.; Albuquerque, S.D.; Cetauro, A.; Lopes, A.; Silva, M.L.D. Dynamics of Heavy Metals in the Waters of Igarape Do Quarenta: The Water Body That Crosses the Industrial Hub in the Brazilian Amazon. Open Sci. J. 2022, 7, 1–13. [Google Scholar]
- Laurance, W.F.; Vasconcelos, H.L.; Lovejoy, T.E. Forest Loss and Fragmentation in the Amazon: Implications for Wildlife Conservation. Oryx 2000, 34, 39–45. [Google Scholar] [CrossRef]
- Feng, X.; Merow, C.; Liu, Z.; Park, D.S.; Roehrdanz, P.R.; Maitner, B.; Newman, E.A.; Boyle, B.L.; Lien, A.; Burger, J.R.; et al. How Deregulation, Drought and Increasing Fire Impact Amazonian Biodiversity. Nature 2021, 597, 516–521. [Google Scholar] [CrossRef]
- Raftopoulos, M.; Morley, J. Ecocide in the Amazon: The Contested Politics of Environmental Rights in Brazil. Int. J. Hum. Rights 2020, 24, 1616–1641. [Google Scholar] [CrossRef]
- De Oliveira, G.M.; Sellare, J.; Börner, J. Mind Your Language: Political Discourse Affects Deforestation in the Brazilian Amazon; ZEF Discussion Papers on Development Policy N. 326; ZEF: Bonn, Germany, 2023. [Google Scholar]
- Zuluaga, S.; Vargas, F.H.; Kohn, S.; Grande, J.M. Top-down Local Management, Perceived Contribution to People, and Actual Detriments Influence a Rampant Human—top Predator Conflict in the Neotropics. Perspect. Ecol. Conserv. 2022, 20, 91–102. [Google Scholar] [CrossRef]
- Nogueira, C.C.; Argôlo, A.J.S.; Arzamendia, V.; Azevedo, J.A.; Barbo, F.E.; Bérnils, R.S.; Bolochio, B.E.; Borges-Martins, M.; Brasil-Godinho, M.; Braz, H.; et al. Atlas of Brazilian Snakes: Verified Point-Locality Maps to Mitigate the Wallacean Shortfall in a Megadiverse Snake Fauna. South Am. J. Herpetol. 2019, 14, 1–274. [Google Scholar] [CrossRef]
- Wagler, J.G. Natürliches System der Amphibien: Mit Vorangehender Classification der Säugethiere und Vögel: Ein Beitrag zur Vergleichenden Zoologie; der J. G. Cotta’schen Buchhandlung: München, Germany, 1830. [Google Scholar]
- Lopez, C.G. Fauna Legendaria; Editorial Arte: Caracas, Venezuela, 1984. [Google Scholar]
- Rivas, J.A. Anaconda: The Secret Life of the Wolrd’s Largest Snake; Oxford University Press: Oxford, UK, 2020. [Google Scholar]
- Tarkhnishvili, D.; Hille, A.; Waller, T.; Todua, M.; Murtskhvaladze, M.; Böhme, W. Morphological Trends and Genetic Divergence in Anacondas, Genus Eunectes Wagler, 1830 (Serpentes: Boidae). Amphibia-Reptilia 2022, 43, 379–393. [Google Scholar] [CrossRef]
- Dirksen, L. Anakondas: Monographische Revision der Gattung Eunectes Wagler, 1830 (Serpentes, Boidae); Natur und Tier—Verlag: Münster, Germany, 2002. [Google Scholar]
- Dirksen, L.; Böhme, W. Studies on Anacondas III. A Reappraisal of Eunectes beniensis Dirksen, 2002, from Bolivia, and a Key to the Species of the Genus Eunectes Wagler, 1830 (Serpentes: Boidae). Russ. J. Herpetol. 2005, 12, 223–229. [Google Scholar]
- Powell, R.L.; Eversole, C.B.; Rivas, L.R.; Crocker, A.V.; De la Quintana, P. Feliz cumpleaños, 21 years for the Beni Anaconda, Eunectes beniensis (Dirksen, 2002) (Serpentes, Boidae): An update of voucher specimens, species’ distribution, and clarification of locality data of type specimens. Check List 2023, 19, 847–854. [Google Scholar] [CrossRef]
- Dirksen, L.; Henderson, R.W. Eunectes deschauenseei Dunn and Conant de Schauensee’s Anacondaz. Cat. Am. Amphib. Reptiles 2002, 755, 1–3. [Google Scholar]
- Dirksen, L.; Oubotar, P. Eunectes deschauenseei. In The IUCN Red List of Threatened Species 2021; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2021. [Google Scholar] [CrossRef]
- Santos, G.S.; de Lema, T.; Winck, G.R.; Cechin, S.Z.; Boelter, R.A. Distribution Extension of the Yellow Anaconda Eunectes notaeus Cope, 1862 (Squamata: Boidae) in the state of Rio Grande do Sul, Brazil. Check List. 2013, 9, 660–662. [Google Scholar] [CrossRef]
- De La Quintana, P.; Rivas, J.A.J.A.; Valdivia, F.; Pacheco, L.F.L.F. Home Range and Habitat Use of Beni Anacondas (Eunectes beniensis) in Bolivia. Amphib. Reptil. 2017, 38, 547–553. [Google Scholar] [CrossRef]
- De la Quintana, P.; Rivas, J.A.; Valdivia, F. Eunectes murinus (Green Anaconda): Dry Season Home Range. Herpetol. Rev. 2018, 49, 546–547. [Google Scholar]
- Smaniotto, N.P.; Moreira, L.F.B.; Rivas, J.A.; Strüssmann, C. Home Range Size, Movement, and Habitat Use of Yellow Anacondas (Eunectes notaeus). Salamandra 2020, 56, 159–167. [Google Scholar]
- Rivas, J.A. Natural History of the Green Anaconda: With Emphasis on Its Reproductive Biology; CreateSpace Independent Publishing Platform: Scotts Valley, CA, USA, 2015. [Google Scholar]
- Rivas, J.A.; del Muñoz, M.C.; Thorbjarnarson, J.B.; Burghardt, G.M.; Holmstrom, W.; Calle, P. Natural History of the Green Anacondas in the Venezuelan Llanos. In Biology of Boas, Pythons, and Related Taxa; Henderson, R.W., Powell, R., Eds.; Eagle Mountain Publishing Company: Eagle Mountain, UT, USA, 2007; pp. 128–138. [Google Scholar]
- Rivas, J.A.; Molina, C.R.; Corey-Rivas, S.J.; Burghardt, G.M. Natural History of Neonatal Green Anacondas (Eunectes murinus): A Chip off the Old Block. Copeia 2016, 104, 402–410. [Google Scholar] [CrossRef]
- Rivas, J.A. Predatory Attacks of Green Anacondas (Eunectes murinus) on Adult Human Beings. Herpetol. Nat. Hist. 1998, 6, 157–159. [Google Scholar]
- Rivas, J.A. Eunectes murinus (Green Anaconda): Subduing Behavior. Herpetol. Rev. 2004, 35, 66–67. [Google Scholar]
- Rivas, J.A.; Ascanio, R.E.; Munoz, M.D. What Is the Length of a Snake? Contemp. Herpetol. 2008, 2008, 1–3. [Google Scholar] [CrossRef]
- Miranda, E.B.P.; Ribeiro-Jr, R.P.; Camera, B.F.; Barros, M.; Draque, J.; Micucci, P.; Waller, T.; Strüssmann, C. Penny and Penny Laid Up Will Be Many: Large Yellow Anacondas Do Not Disregard Small Prey. J. Zool. 2017, 301, 301–309. [Google Scholar] [CrossRef]
- Strüssman, C. Hábitos Alimantares Da Sucuri-Amarela, Eunectes notaeus Cope, 1862, No Pantanal Matogrossense. Biociencias 1997, 1, 35–52. [Google Scholar]
- Valderrama, X.; Thorbjarnarson, J.B. Eunectes murinus: Predation. Herpetol. Rev. 2001, 32, 46–47. [Google Scholar]
- Rivas, J.A.; Thorbjarnarson, J.B.; Owens, R.Y.; Muñoz, M.d.C. Eunectes murinus: Caiman Predation. Herpetol. Rev. 1999, 30, 101. [Google Scholar]
- Rivas, J.A.; Owens, R.Y.; Calle, P.P. Eunectes murinus: Juvenile Predation. Herpetol. Rev. 2001, 32, 107–108. [Google Scholar]
- Terra, J.D.S. Ecologia, Nicho Climático e Efeito Das Mudanças Climáticas Sobre a Distribuição Potencial Das Espécies Do Gênero Eunectes (Squamata, Serpente). Ph.D. Thesis, Universidade de São Paulo, Sao Paulo, Brazil, 2018. [Google Scholar]
- Champagne, P. Stephane. Conservation Ecology of Eunectes murinus (Green Anaconda) in the Madre de Dios Region of Southeastern Peru Using Remote Sensing Techniques and Machine Learning Driven Geospatial Modeling. Ph.D. Thesis, Acadia University, Wolfville, NS, Canada, 2022. [Google Scholar]
- Calle, P.P.; Rivas, J.A.; Munoz, M.d.C.; Thorbjarnarson, J.; Dierenfeld, E.S.; Holmstrom, W.; Karesh, W.E.B.W.B. Health Assessment of Free Ranging Anacondas (Eunectes murinus) in Venezuela. J. Zoo Wildl. Med. 1994, 25, 53–61. [Google Scholar]
- Calle, P.P.P.; Rivas, J.A.; Muñoz, M.; Thorbjarnarson, J.B.; Holmstrom, W.; Karesh, W.B.W.B. Infectious Disease Serologic Survey in Free-Ranging Venezuelan Anacondas (Eunectes murinus). J. Zoo Wildl. Med. 2001, 32, 320–323. [Google Scholar] [CrossRef]
- Rivas, J.A. Determining Breeding Status of Green Anacondas (Eunectes murinus): A Condition Index Assuming Isometry. South Am. J. Herpetol. 2023, 28, 89–94. [Google Scholar] [CrossRef]
- Rivas, J.A. What Can Studying Anacondas Tell Us about Titanoboa cerrejonensis? Exploring the Life of an Extinct Giant Snake Using an Extant Pretty Big Snake. Herpetol. J. 2023, 33, 68–75. [Google Scholar] [CrossRef]
- Rivas, J.A.; Corey-Rivas, S.J. Eunectes murinus (Green Anaconda), Longevity. Herpetol. Rev. 2008, 39, 469. [Google Scholar]
- Rivas, J.A. Conservation of Green Anacondas: How Tylenol Conservation and Macroeconomics Threaten the Survival of the World’s Largest Snake. Iguana 2007, 14, 10–21. [Google Scholar]
- Rivas, J.A.; del Munoz, M.C.; Thorbjarnarson, J.B.; Burghardt, G.M. Eunectes murinus. In Boas of the World; Reynolds, R.G., Robert, W.H., Eds.; In Revision; Comstock Publishing Associates: Ithaca, NY, USA.
- Calderón, M.; Ortega, A.; Scott, N.; Cacciali, P.; de Nogueira, C.C.; Gagliardi, G.; Catenazzi, A.; Cisneros-Heredia, D.F.; Hoogmoed, M.S.; Schargel, W.; et al. Eunectes murinus. In The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2001; Available online: https://www.iucnredlist.org/species/44580041/44580052 (accessed on 15 February 2024).
- Waller, T.; Micucci, P.A.; Alvarenga, E. Conservation Biology of the Yellow Anaconda (Eunectes notaeus) in Northeastern Argentina. In Biology of Boas, Pythons, and Related Taxa; Henderson, R.W., Powell, R., Eds.; Eagle Mountain Publishing Company: Eagle Mountain, UT, USA, 2007; pp. 340–362. [Google Scholar]
- Micucci, P.A.; Waller, T.; Alvarenga, E. Programa Curiyú. In Manejo de Fauna Silvestre en la Argentina: Programa de Uso Sustentable; Bolkovic, M.L., Ramadori, D., Eds.; Ministerio de Salud y Ambiente de la Nación: Buenos Aires, Argentina, 2006; pp. 77–92. [Google Scholar]
- Waller, T.; Micucci, P.A. Anaconda Conservation: A Reply to Rivas. Iguana 2008, 15, 51–53. [Google Scholar]
- Rivas, J.A. Is Wildlife Management Business or Conservation—A Question of Ideology. Reptiles Amphib. 2010, 17, 112–115. [Google Scholar] [CrossRef]
- Camera, B.F.; Quintana, I.; Strussmann, C.; Waller, T.; Barros, M.; Draque, J.; Micucci, P.A.; Miranda, E.B.P. Assessing the Sustainability of Yellow Anaconda (Eunectes notaeus) Harvest. PLoS ONE 2023, 18, e0277629. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.R.N.; Pereira Filho, G.A. Commercialization and Use of Snakes in North and Northeastern Brazil: Implications for Conservation and Management. Biodivers. Conserv. 2007, 16, 969–985. [Google Scholar] [CrossRef]
- Giraudo, A.R.; Arzamendia, V.; Bellini, G.P.; Bessa, C.A.; Calamante, C.C.; Cardozo, G.; Chiaraviglio, M.; Constanzo, M.B.; Etchepare, E.G.; Di Cola, V.; et al. Categorización Del Estado de Conservación de Las Serpientes de La República Argentina. Cuad. Herpetol. 2012, 26, 303–326. [Google Scholar]
- Waller, T.; Camera, B.; Cacciali, P.; Miranda, E.; Buongermini, E.; Micucci, P.; Smaniotto, N.; Strüssmann, C.; Barros, M.; Draque, J. Eunectes notaeus. In The IUCN Red List of Threatened Species; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2021. [Google Scholar] [CrossRef]
- Carreira, S.; Estrades, A. Reptil. In Especies Prioritarias para la Conservación en Uruguay: Vertebrados, Moluscos Continentales y Plantas Vasculares; Soutullo, A., Clavijo, C., Martínez-Lanfranco, J.A., Eds.; SNAP/DINAMA/MVOTMA y DICYT/MEC: Montevideo, Uruguay, 2013; pp. 129–147. [Google Scholar]
- Micucci, P.A.; Waller, T. The Management of Yellow Anacondas (Eunectes notaeus) in Argentina: From Historical Misuse to Resource Appreciation. Iguana 2007, 14, 160–171. [Google Scholar]
- Muñoz, A.; Gonzales, L.; Embert, D.; Aparicio, J.; Aguayo, R. Eunectes beniensis . Available online: https://www.iucnredlist.org/species/174126/18978378 (accessed on 15 February 2024).
- Rivas, J.A. ¿Cuantas Anacondas Hay? In Proceedings of the XI Latin American Congress of Herpetology, Quito, Ecuador, 24–28 July 2017. [Google Scholar]
- Rivas, J.A.; Corey-Rivas, S.J. How Many Green Anacondas Are There?: Evidence of Multiple Lineages of Green Anaconda. In Proceedings of the Joint Meeting of Ichthyologist and Herpetologist 2017, Austin, TX, USA, 12–17 July 2017. [Google Scholar]
- Seutin, G.; White, B.N.; Boag, P.T. Preservation of Avian Blood and Tissue Samples for DNA Analyses. Can. J. Zool. 1991, 69, 82–90. [Google Scholar] [CrossRef]
- Douglas, D.A.; Janke, A.; Arnason, U. A Mitogenomic Study on the Phylogenetic Position of Snakes. Zool. Scr. 2006, 35, 545–558. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- de Queiroz, A.; Lawson, R.; Lemos-Espinal, J. A Phylogenetic Relationships of North American Garter Snakes (Thamnophis) Based on Four Mitochondrial Genes: How Much DNA Sequence Is Enough? Mol. Phylogenet. Evol. 2002, 22, 315–329. [Google Scholar] [CrossRef]
- Burbrink, F.T.; Lawson, R.; Slowinski, J.B. Mitochondrial DNA Phylogeography of the Polytypic North American Rat Snake (Elaphe Obsoleta): A Critique of the Subspecies Concept. Evolution 2000, 54, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, E.; Davis, S.K.; Sites, J.W. Mitochondrial DNA Sequence Divergence and Phylogenetic Relationships among Eight Chromosome Races of the Sceloporus grammicus Complex (Phrynosomatidae) in Central Mexico. Soc. Syst. Biol. 1994, 43, 387–418. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Noonan, B.P.; Chippindale, P.T. Dispersal and Vicariance: The Complex Evolutionary History of Boid Snakes. Mol. Phylogenet. Evol. 2006, 40, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Diaz, J. Identification of Single Copy Nuclear DNA Markers for North American Pit Vipers. Mol. Ecol. Resour. 2010, 10, 177–180. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. RaxmlGUI 2.0: A Graphical Interface and Toolkit for Phylogenetic Analyses Using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.M.; Teslenko, P.; van der Mark, D.L.; Ayres, A.; Darling, S.; Höhna, B.; Larget, L.; Liu, M.A.S.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian Phylo-Genetic Inference and Model Selection across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Smith, K.T.; Scanferla, A. A Nearly Complete Skeleton of the Oldest Definitive Erycine Boid (Messel, Germany). Geodiversitas 2021, 43, 1–24. [Google Scholar] [CrossRef]
- Head, J.J. Fossil Calibration Dates for Molecular Phylogenetic Analysis of Snakes 1: Serpentes, Alethinophidia, Boidae, Pythonidae. Palaeontol. Electron. 2015, 18, 1–17. [Google Scholar] [CrossRef]
- Storey, M.; Mahoney, J.J.; Saunders, A.D.; Duncan, R.A.; Kelley, S.P.; Coffin, M.F. Timing of Hot Spot—Related Volcanism and the Breakup of Madagascar and India. Science 1995, 267, 852–855. [Google Scholar] [CrossRef]
- Ali, J.R.; Krause, D.W. Late Cretaceous Bioconnections between Indo-Madagascar and Antarctica: Refutation of the Gunnerus Ridge Causeway Hypothesis. J. Biogeogr. 2011, 38, 1855–1872. [Google Scholar] [CrossRef]
- Prance, G.T. Biological Diversification in the Tropics; Columbia University Press: New York, NY, USA, 1982. [Google Scholar]
- Junk, W.J.; Wittmann, F.; Schöngart, J.; Piedade, M.T.F. A Classification of the Major Habitats of Amazonian Black-Water River Floodplains and a Comparison with Their White-Water Counterparts. Wetl. Ecol. Manag. 2015, 23, 677–693. [Google Scholar] [CrossRef]
- Rivas, J.A. The Missing River. Front. Earth Sci. 2023, 11, 1203667. [Google Scholar] [CrossRef]
- Roze, J.A. La Taxonomía y Zoogeografía de Los Ofidios de Venezuela; Universidad Central de Venezuela, Ediciones de la Biblioteca: Caracas, Venezuela, 1966. [Google Scholar]
- Gorzula, S.; Pilgrim, K. Snakes of the Family Boidae in the Cooperative Republic of Guayana: An Assessment of the Trade and Status of Sixe Species; Report to the Secretariat of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES): Lausanne, Switzerland, 1992. [Google Scholar]
- Wiens, J.J.; Tiu, J. Highly Incomplete Taxa Can Rescue Phylogenetic Analyses from the Negative Impacts of Limited Taxon Sampling. PLoS ONE 2012, 7, e42925. [Google Scholar] [CrossRef] [PubMed]
- von Linnaeus, C. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis; Salvii, L., Ed.; Impensis Direct: Holmiae, Sweden, 1758; Volume 1. [Google Scholar]
- Gronovius, L.T. Museum Ichthyologicum, Sistens Piscium Indigenorum & Quorumdam Exoticorum; Haak, T., Ed.; Lugduni-Batavorum: Leiden, The Netherlands, 1756; Volume 3. [Google Scholar] [CrossRef]
- Dirksen, L.; Bohme, W. Studien an Anakondas. I: Indizien Filr Natiirliche Bastardierung Zwischen Der Groben Anakonda Eunectes murinus Und Der Paraguayanakon da Eunectes notaeus in Bolivien, Mit Bemerkungen Zur Taxonomie der Gattung Eunectes. Zool. Abh. Staatl. Mus. Dresden 1998, 4, 45–58. [Google Scholar]
- Turci, L.C.B.; Bernarde, P.S. Composition and Natural History of Snakes from Zona Da Mata in Rondônia, Southwestern Brazilian Amazon. J. Neotrop. Biol. 2022, 19, 111–126. [Google Scholar] [CrossRef]
- Shankar, P.G.; Swamy, P.; Williams, R.C.; Ganesh, S.R.; Moss, M.; Höglund, J.; Das, I.; Sahoo, G.; Vijayakumar, S.P.; Shanker, K.; et al. King or Royal Family? Testing for Species Boundaries in the King Cobra, Ophiophagus hannah (Cantor, 1836), Using Morphology and Multilocus DNA Analyses. Mol. Phylogenet. Evol. 2021, 165, 107300. [Google Scholar] [CrossRef]
- Brandt, A.L.; Ishida, Y.; Georgiadis, N.J.; Roca, A.L. Forest Elephant Mitochondrial Genomes Reveal That Elephantid Diversification in Africa Tracked Climate Transitions. Mol. Ecol. 2012, 21, 1175–1189. [Google Scholar] [CrossRef]
- Murphy, J.C.; Jowers, M.J.; Lehtinen, R.M.; Charles, S.P.; Colli, G.R.; Peres, A.K.; Hendry, C.R.; Pyron, R.A. Cryptic, Sympatric Diversity in Tegu Lizards of the Tupinambis teguixin Group (Squamata, Sauria, Teiidae) and the Description of Three New Species. PLoS ONE 2016, 11, e0158542. [Google Scholar] [CrossRef]
- Avila, I.; Bauer, F.; Boungermini, E.; Martinez, N. Dracaena paraguayensis: Aportes Sobre El Conocimiento De Su Biología, Ecología y distribución. Kempffiana 2016, 12, 122–130. [Google Scholar]
- Avila-pires, T.C.S. Lizards of Brazilian Amazonia (Reptilia: Squamata). Zool. Verh. 1995, 599, 1–706. [Google Scholar]
- Vargas-Ramírez, M.; Caballero, S.; Morales-Betancourt, M.A.; Lasso, C.A.; Amaya, L.; Martínez, J.G.; das Neves Silva Viana, M.; Vogt, R.C.; Farias, I.P.; Hrbek, T.; et al. Genomic Analyses Reveal Two Species of the Matamata (Testudines: Chelidae: Chelus spp.) and Clarify Their Phylogeography. Mol. Phylogenet. Evol. 2020, 148, 106823. [Google Scholar] [CrossRef] [PubMed]
- Michels, J.; Vargas-Ramírez, M. Red-Headed Amazon River Turtles in Venezuela and Colombia: Population Separation and Connection along the Famous Route of Alexander von Humboldt. Zoology 2018, 130, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; Henderson, R.W.; Delmas, A.-S.S.; Hedges, S.B. A Phylogenetic Study of the Emerald Treeboa (Corallus caninus). J. Herpetol. 2005, 39, 500–503. [Google Scholar] [CrossRef]
- Colston, T.J.; Grazziotin, F.G.; Shepard, D.B.; Vitt, L.J.; Colli, G.R.; Henderson, R.W.; Hedges, B.; Zaher, H.; Burbrink, F.T. Molecular Systematics and Historical Biogeography of Tree Boas (Corallus spp.). Mol. Phylogenet. Evol. 2013, 66, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Passos, P.; Fernandes, R. Revision of the Epicrates cenchria Complex (Serpentes: Boidae). Herpetol. Monogr. 2008, 22, 1–30. [Google Scholar] [CrossRef]
- Seba, A. Locupletissimi Rerum Naturalium Thesauri Accurata Descriptio, et Iconibus Artificiosissimis Expressio, per Universam Physices Historiam: Opus, Cui, in Hoc Rerum Genere, Nullum Par Exstitit. Tomus II; Wetstenium, J., Smith, G., Waesberg, J., Eds.; Amstelaedami: Amsterdam, The Netherlands, 1735; ISBN 9788527729833. [Google Scholar]
- Bauer, A.M.; Wahlgren, R. On the Linck Collection and Specimens of Snakes Figured by Johann Jakob Scheuchzer (1735)—The Oldest Fluid-Preserved Herpetological Collection in the World? Bonn Zool. Bull. 2013, 62, 220–252. [Google Scholar]
- Scheuchzer, J.J. Physica Sacra; Wagner, C.U., Ed.; Christian Ulrich Wagner: Augsburg and Ulm, Germany, 1735; Volume 4, ISBN 2013206534. [Google Scholar]
- Plotkin, M.J. Tales of a Shaman’s Apprentice: An Ethnobotanist Searches for New Medicines in the Rain Forest; Penguin: New York, NY, USA, 1994. [Google Scholar]
- Gumilla, J. El Orinoco Ilustrado y Defendido; Editorial Arte: Madrid/Caracas, Spain, 1740. [Google Scholar]
- Dunn, E.R.; Conant, R. Notes on Anacondas, with Descriptions of Two New Species. Proc. Acad. Nat. Sci. Phila. 1936, 88, 503–506. [Google Scholar]
- Dirksen, L.; Bohme, W. Studien an Anakondas. II: Zurn Taxonomischen Status von Eunectes murinus gigas (Latreille, 1801) (Serpentes: Boidae), Mit Neuen Ergebnis-Sen Zur Gattung Eunectes Wagler, 1830. Salamandra 1998, 34, 359–374. [Google Scholar]
- International Commission on Zoological Nomenclature. International Code of Zoological Nomenclature, 4th ed.; International Trust for Zoological Nomenclature: London, UK, 1999. [Google Scholar]
- Reynolds, R.G.; Niemiller, M.L.; Hedges, S.B.; Dornburg, A.; Puente-Rolón, A.R.; Revell, L.J. Molecular Phylogeny and Historical Biogeography of West Indian Boid Snakes (Chilabothrus). Mol. Phylogenet. Evol. 2013, 68, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.G.; Marshall, L.G.; Guerrero, J.; Horton, B.; Malabarba, M.C.S.L.; Wesselingh, F. The Stage for Neotropical Fish Diversification: A History of Tropical South American Rivers. In Phylogeny and Classification of Neotropical Fishes; Malabarba, L.R., Reis, R.E., Vari, R.P., Lucena, Z.M.S., Lucena, C.A.S., Eds.; Edipucrs: Porto Alegre, Brazil, 1998; pp. 13–48. [Google Scholar]
- Bicudo, T.C.; Sacek, V.; de Almeida, R.P.; Bates, J.M.; Ribas, C.C. Andean Tectonics and Mantle Dynamics as a Pervasive Influence on Amazonian Ecosystem. Sci. Rep. 2019, 9, 16879. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, X.; Nie, L.; Xia, X.; Liu, L.; Jiang, Y.; Huang, Z.; Jing, W. The Complete Mitochondrial Genome Sequences of Chelodina rugosa and Chelus fimbriata (Pleurodira: Chelidae): Implications of a Common Absence of Initiation Sites (OL) in Pleurodiran Turtles. Mol. Biol. Rep. 2012, 39, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, G.S.; Bronzati, M.; Langer, M.C.; Sterli, J. Phylogeny, Biogeography and Diversification Patterns of Side-Necked Turtles (Testudines: Pleurodira). R. Soc. Open Sci. 2018, 5, 171773. [Google Scholar] [CrossRef] [PubMed]
- Roberto, I.J.; Bittencourt, P.S.; Muniz, F.L.; Hernández-Rangel, S.M.; Nóbrega, Y.C.; Ávila, R.W.; Souza, B.C.; Alvarez, G.; Miranda-Chumacero, G.; Campos, Z.; et al. Unexpected but Unsurprising Lineage Diversity within the Most Widespread Neotropical Crocodilian Genus Caiman (Crocodylia, Alligatoridae). Syst. Biodivers. 2020, 18, 377–395. [Google Scholar] [CrossRef]
- Villamarín, F.; Escobedo-Galván, A.H.; Siroski, P.; Magnusson, W.E. Geographic Distribution, Habitat, Reproduction, and Conservation Status of Crocodilians in the Americas. In Conservation Genetics of New World Crocodilians; Zucoloto, R.B., Amavet, P.S., Verdade, L.M., Farias, I.P., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Giugliano, L.G.; Collevatti, R.G.; Colli, G.R. Molecular Dating and Phylogenetic Relationships among Teiidae (Squamata) Inferred by Molecular and Morphological Data. Mol. Phylogenet. Evol. 2007, 45, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Waller, T.; Buongermini, E.; Micucci, P.A. Eunectes notaeus. Diet. Herpetol. Rev. 2001, 32, 47. [Google Scholar]
- Crawford, R.M.M. Oxygen availability as an ecological limit to plant distribution. In Advances in Ecological Research; Begon, M., Fitter, A.H., Eds.; Academic Press: Cambridge, MA, USA, 1992; Volume 23, pp. 93–185. [Google Scholar]
- Wittmann, F.; Householder, J.E.; Piedade, M.T.F.; Schöngart, J.; Demarchi, L.O.; Quaresma, A.C.; Junk, W.J. A Review of the Ecological and Biogeographic Differences of Amazonian Floodplain Forests. Water 2022, 14, 3360. [Google Scholar] [CrossRef]
- Hoorn, C.; Wesselingh, F.P.; Steege, H.T.; Bermudez, M.A.; Mora, A.; Sevink, J.; Sanmartín, I.; Sanchez-Meseguer, A.; Anderson, C.L.; Figueiredo, J.P.; et al. Amazonia Through Time: Andean Uplift, Climate Change, Landscape Evolution, and Biodiversity. Science 2010, 330, 927. [Google Scholar] [CrossRef]
- Wesselingh, F.P.; Rasanen, M.E.; Irion, G.; Vonhof, H.G.; Kaandorp, R.J.G.; Renema, W.; Romero-Pittman, L.; Gingras, M. Lake Pebas: Palaeoecological Reconstruction of a Miocene Long-Lived Lake Complex in Western Amazonia. Cainozoic Res. 2002, 2001, 35–81. [Google Scholar]
- Hoorn, C.; Guerrero, J.; Sarmiento, G.; Lorente, M.A. Andean Tectonics as a Cause for Changing Drainage Patterns in Miocene Northern South America. Geology 1995, 23, 237–240. [Google Scholar] [CrossRef]
- Winemiller, K.O.; Willis, S.C. The Vaupes Arch and Casiquiare Canal: Barriers and Passages; Albert, J.S., Reis, R.E., Eds.; University of California Press: Berkeley, CA, USA, 2011; ISBN 0520268685. [Google Scholar]
- Rivas, J.A.; Rodriguez, J.V.; Mittermeier, C.G. The Llanos. In Wilderness: Earth’s Last Wildl Places; Mittermeier, R., Mittermeier, C.G., Robles Gil, P., Pilgrim, J., Konstant, W.R., Fonseca, G.A.B., Brooks, T.M., Eds.; Cemex: San Pedro Garza García, Mexico, 2002; Volume 53, pp. 1689–1699. ISBN 9788578110796. [Google Scholar]
- Sarmiento, G. The Savannas of Tropical America. In Ecosystems of the World: Tropical Savannas; Bourliere, F., Ed.; Elsevier Scientifc Publishing Co.: New York, NY, USA, 1983; Volume 13, pp. 245–288. [Google Scholar]
- Hoorn, C.; Boschman, L.M.; Kukla, T.; Sciumbata, M.; Val, P. The Miocene Wetland of Western Amazonia and Its Role in Neotropical Biogeography. Bot. J. Linn. Soc. 2022, 199, boab098. [Google Scholar] [CrossRef]
- Antoine, P.O.; Abello, M.A.; Adnet, S.; Altamirano Sierra, A.J.; Baby, P.; Billet, G.; Boivin, M.; Calderón, Y.; Candela, A.; Chabain, J.; et al. A 60-Million-Year Cenozoic History of Western Amazonian Ecosystems in Contamana, Eastern Peru. Gondwana Res. 2016, 31, 30–59. [Google Scholar] [CrossRef]
- Salas-Gismondi, R.; Flynn, J.J.; Baby, P.; Tejada-Lara, J.V.; Wesselingh, F.P.; Antoine, P.O. A Miocene Hyperdiverse Crocodylian Community Reveals Peculiar Trophic Dynamics in Proto-Amazonian Mega-Wetlands. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142490. [Google Scholar] [CrossRef] [PubMed]
- Tucker, D.B. Molecular Studies of South American Teiid Lizards (Teiidae: Squamata) from Deep Time to Shallow Divergences from Deep Time to Shallow Divergences. Ph.D. Thesis, Brigham University, Provo, UT, USA, 2016. [Google Scholar]
- Kirschner, J.A.; Hoorn, C. The Onset of Grasses in the Amazon Drainage Basin, Evidence from the Fossil Record. Front. Biogeogr. 2020, 12, e44827. [Google Scholar] [CrossRef]
- Leite, Y.L.R.; Costa, L.P.; Loss, A.C.; Rocha, R.G.; Batalha-Filho, H.; Bastos, A.C.; Quaresma, V.S.; Fagundes, V.; Paresque, R.; Passamani, M.; et al. Neotropical Forest Expansion during the Last Glacial Period Challenges Refuge Hypothesis. Proc. Natl. Acad. Sci. USA 2016, 113, 1008–1013. [Google Scholar] [CrossRef]
- Natsukawa, H.; Sergio, F. Top Predators as Biodiversity Indicators: A Meta-Analysis. Ecol. Lett. 2022, 25, 2062–2075. [Google Scholar] [CrossRef]
- Sergio, F.; Newton, I.; Marchesi, L. Top Predators and Biodiversity. Nature 2005, 436, 192. [Google Scholar] [CrossRef]
- Ojasti, J. Manejo de Fauna Silvestre Neotropical; Dallmeier, F., Ed.; Smithsonian Institution Press: Washington, DC, USA, 2000; ISBN 189391206X. [Google Scholar]




| Gene and Primer | Primer Sequences | Mt Location and References |
|---|---|---|
| Cytochrome b | ||
| L14910_cytbEu | 5′-GACCTGMGGTCTGAAAAACCACCG TTG T-3′ | tRNA-Glu, [70,71], modified by this study |
| H16064_cytbEu | 5′-CTTTGGTTTACAGAACAATGCTTTG-3′ | tRNA-Thr, [71], modified by this study |
| ND2 | ||
| En5132F | 5′-AATGTCACCACGGCCTTTAC-3′ | tRNA-met, this study |
| En6262R | 5′-TGCAGGCTCTACAGAAGCTAAA-3′ | tRNA-Trp/tRNA-Ala, this study |
| Eu_ND2_6003 | 5′-TGGCTATTGTYGTKGCTTCT-3′ | ND2, this study |
| ND4 | ||
| ND4_Eu | 5′-CACCTATGACTACCAAAAGCCCACGTAGAAGC-3′ | ND4, [72], modified by this study |
| ND4_LEU | 5′-CATTTCTRCYACTTGGATTTGCACCA-3′ | tRNA-Leu, Arévalo et al., 1994, modified by this study |
| Approach | Calibration Point Type | Node | Prior Implementation in BEAST (v.2.7.6.) | Minimum | Maximum |
|---|---|---|---|---|---|
| 1. Divergence of Sanziniinae from other Boidae assuming possible dispersal to Madagascar via two Late Cretaceous land bridges (Gunnerus Ridge/Kerguelen Plateau). | Paleogeographic and fossil. | LCA of Sanziniinae and other boids. | Log-normal. With hard minimum and soft maximum (represented by 95% percentile). | 80 Million Years Ago (Mya) based on the latest possible existence of Gunnerus Ridge [83]. | 98.32 Mya as soft maximum, based on the fossil Coniophis, the oldest crown-group fossil for Serpentes [79]. |
| Fossil. | LCA of Erycinae and Boinae. | Uniform with only hard minimum. | 58 Mya based on Titanoboa cerrejonensis [79]. | n.a. | |
| Fossil. | LCA of Lichanura and Charina. | Uniform with only hard minimum. | 18.7 Mya based on the fossil UNSM 125,562 [79]. | n.a. | |
| Fossil. | LCA of Corallus and its sister lineage. | Uniform with only hard minimum. | 50.2 Mya based on the fossil Corallus priscus [79]. | n.a. | |
| Fossil. | LCA of Eunectes and Epicrates. | Uniform with only hard minimum. | 12.375 Mya based on Eunectes stirtoni [79]. | n.a. | |
| Fossil. | LCA of Charininae and Ungaliophiinae. | Uniform with only hard minimum. | 47.8 Mya based on the fossil Rageryx schmidi [81]. | n.a. | |
| 2. Divergence of Sanziniinae from the rest of Boidae assuming possible dispersal to Madagascar via the Kerguelen Plateau and the Indian subcontinent. | Paleogeographic and fossil. | Log-normal. With hard minimum and soft maximum (represented by 95% percentile). | 88 Mya based on the latest possible terrestrial connection between Madagascar and the Indian subcontinent [83]. | 98.32 Mya as soft maximum, based on the fossil Coniophis, the oldest crown-group fossil for Serpentes [79]. | |
| Fossil. | LCA of Erycinae and Boinae. | Uniform with only hard minimum. | 58 Mya based on Titanoboa cerrejonensis [79]. | n.a. | |
| Fossil. | LCA of Lichanura and Charina. | Uniform with only hard minimum. | 18.7 Mya based on the fossil UNSM 125,562 [79]. | n.a. | |
| Fossil. | LCA of Corallus and its sister lineage. | Uniform with only hard minimum. | 50.2 Mya based on the fossil Corallus priscus [79]. | n.a. | |
| Fossil. | LCA of Eunectes and Epicrates. | Uniform with only hard minimum. | 12.375 Mya based on Eunectes stirtoni [79]. | n.a. | |
| Fossil. | LCA of Charininae and Ungaliophiinae. | Uniform with only hard minimum. | 47.8 Mya based on the fossil Rageryx schmidi [81]. | n.a. | |
| 3. Divergence of Sanziniinae from the rest of Boidae in the absence of Late Cretaceous land bridges connecting Madagascar to western Gondwana. | Fossil and paleogeographic. | Log-normal. With hard minimum and soft maximum (represented by 95% percentile). | 120.4 Mya based on the split between eastern and western Gondwana (Krause et al. 2020). | 145 Mya based on the oldest fossils testifying for the split between snakes and Anguimorph lizards [79]. | |
| Fossil. | LCA of Erycinae and Boinae. | Uniform with only hard minimum. | 58 Mya based on Titanoboa cerrejonensis [79]. | n.a. | |
| Fossil. | LCA of Lichanura and Charina. | Uniform with only hard minimum. | 18.7 Mya based on the fossil UNSM 125,562 [79]. | n.a. | |
| Fossil. | LCA of Corallus and its sister lineage. | Uniform with only hard minimum. | 50.2 Mya based on the fossil Corallus priscus [79]. | n.a. | |
| Fossil. | LCA of Eunectes and Epicrates. | Uniform with only hard minimum. | 12.375 Mya based on Eunectes stirtoni [79]. | n.a. | |
| Fossil. | LCA of Charininae and Ungaliophiinae. | Uniform with only hard minimum. | 47.8 Mya based on the fossil Rageryx schmidi [81]. | n.a. | |
| 4. Strictly paleontological calibration based on [82]. | Fossil. | Pan-serpentes crown-group age based on Coniophis. | Log-normal. With hard minimum and soft maximum (represented by 95% percentile). | 98.32 Mya | 113 Mya |
| Fossil. | LCA of Erycinae and Boinae based on Titanoboa cerrejonensis. | Log-normal. With hard minimum and soft maximum (represented by 95% percentile). | 58 Mya | 64 Mya | |
| Fossil. | LCA of Corallus and its sister lineage based on Corallus priscus. | Log-normal. With hard minimum and soft maximum (represented by 95% percentile). | 50.2 Mya | 64 Mya | |
| Fossil. | LCA of Charininae and Ungaliophiinae. | Uniform with only hard minimum. | 35.2 Mya | n.a. | |
| Fossil. | LCA of Charina and Lichanura based on Calamagras weigeli. | Uniform with only hard minimum. | 18.7 Mya | n.a. | |
| Fossil. | LCA of Eunectes and Epicrates. | Uniform with only hard minimum. | 12.375 Mya | n.a. |
| E. murinus | E. akayima | E. notaeus 1 | E. beniensis | E. deschauenseei | |
|---|---|---|---|---|---|
| E. akayima | 5.50% | ||||
| E. notaeus 1 | 11.27% | 10.37% | |||
| E. beniensis | 10.98% | 10.95% | 2.25% | ||
| E. deschauenseei | 10.58% | 10.18% | 0.67% | 2.14% | |
| E. notaeus 2 | 10.94% | 9.91% | 0.74% | 2.38% | 0.81% |
| E. akayima This Study | E. akayima Dirksen (2002) | E. murinus Dirksen (2002) | Linnaeus 1758 (NRM-9) | |
|---|---|---|---|---|
| Ventral | 247–249 | 243–259 | 254 | |
| Subcaudal | 67–68 | 61–69 | 61–78 | 65–69 |
| Dorsal scales, mid-body | 65–65 | 59–66 | 58–74 | |
| Dorsal blotches | 80–90 | 104–116 | 81–148 | 113 |
| Spots in contact | 10–20 | |||
| Supralabials | 14–15 | 16–17 | 14–18 | damaged |
| Infralabials | 16–20 | 20–21 | 18–25 | 16 |
| Infraoculars | 2–3 | 3 | ||
| In contact with eye | 8–9 | 6–8 | 5–10 | damaged |
| Loreal | 4–5 | 3–9 | 3–9 | damaged |
| Supraocular | 1–1 | 1 |
| Scenario | Epicrates - Eunectes Node | Basal Eunectes Genus Node | LCA of E. akayima and E. murinus | LCA of Yellow Anaconda Node |
|---|---|---|---|---|
| Two land bridges (Gunnerus Ridge/Kerguelen Plateau) | 37.68 (51.28–24.01) Mya | 20.81 (31.93–11.18) Mya | 8.70 (15.06–3.95) Mya | 2.85 (5.21–1.31) Mya |
| One land bridge (Kerguelen Plateau only) | 38.62 (52.47–25.10) Mya | 21.54 (33.15–11.68) Mya | 9.08 (15.57–4.44) Mya | 2.96 (5.45–1.38) Mya |
| No land bridges | 46.30 (66.58–28.40) Mya | 26.30 (40.97–14.67) Mya | 11.30 (19.46–5.48) Mya | 3.87 (7.14–1.75) Mya |
| Fossil calibration only [82] | 35.34 (45.41–23.98) Mya | 19.88 (29.61–11.24) Mya | 8.59 (14.43–4.08) Mya | 2.87 (5.28–1.32) Mya |
| Eunectes akayima (MCNG 1042) Holotype 1 | Eunectes akayima (RMNH.RENA.20768) Paratype 2 | Eunectes murinus (MPEG 27428) Lectotype 3 | |
|---|---|---|---|
| Ventral | 241 | 252 | 254 |
| Subcaudal | 53 | 45 | 66 |
| Dorsal scales, anterior body | 50 | 45 | 50 |
| Dorsal scales, mid-body | 60 | 61 | 58 |
| Dorsal, posterior | 41 | 38 | 42 |
| Dorsal spots | 94 | 96 | 94 |
| Spots in contact | 19 | 17 | 18 |
| Supralabials | 14/15 | 16/16 | 16/16 |
| Infralabials | damaged | 22 left side, right side is damaged | 22/22 |
| Infraoculars | damaged | 2 | 2/2 |
| In contact with eye | 7/8 | 7 | 7/7 |
| Suborbitalia | damaged | 4 | 5/5 |
| Supraocular | 1/1 | 1 | 1/1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivas, J.A.; De La Quintana, P.; Mancuso, M.; Pacheco, L.F.; Rivas, G.A.; Mariotto, S.; Salazar-Valenzuela, D.; Baihua, M.T.; Baihua, P.; Burghardt, G.M.; et al. Disentangling the Anacondas: Revealing a New Green Species and Rethinking Yellows. Diversity 2024, 16, 127. https://doi.org/10.3390/d16020127
Rivas JA, De La Quintana P, Mancuso M, Pacheco LF, Rivas GA, Mariotto S, Salazar-Valenzuela D, Baihua MT, Baihua P, Burghardt GM, et al. Disentangling the Anacondas: Revealing a New Green Species and Rethinking Yellows. Diversity. 2024; 16(2):127. https://doi.org/10.3390/d16020127
Chicago/Turabian StyleRivas, Jesús A., Paola De La Quintana, Marco Mancuso, Luis F. Pacheco, Gilson A. Rivas, Sandra Mariotto, David Salazar-Valenzuela, Marcelo Tepeña Baihua, Penti Baihua, Gordon M. Burghardt, and et al. 2024. "Disentangling the Anacondas: Revealing a New Green Species and Rethinking Yellows" Diversity 16, no. 2: 127. https://doi.org/10.3390/d16020127
APA StyleRivas, J. A., De La Quintana, P., Mancuso, M., Pacheco, L. F., Rivas, G. A., Mariotto, S., Salazar-Valenzuela, D., Baihua, M. T., Baihua, P., Burghardt, G. M., Vonk, F. J., Hernandez, E., García-Pérez, J. E., Fry, B. G., & Corey-Rivas, S. (2024). Disentangling the Anacondas: Revealing a New Green Species and Rethinking Yellows. Diversity, 16(2), 127. https://doi.org/10.3390/d16020127