Design of Flow Systems for Improved Networking and Reduced Noise in Biomolecular Signal Processing in Biocomputing and Biosensing Applications
Abstract
:1. Introduction
2. Experimental Setup
2.1. Materials and Reagents
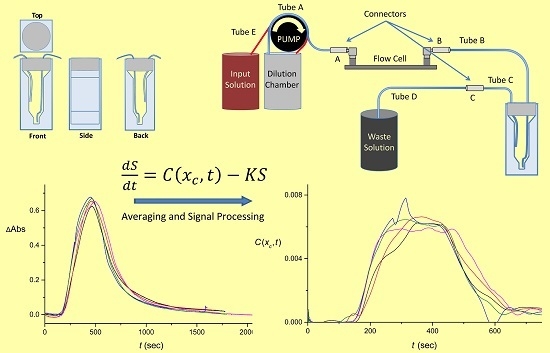
2.2. Flow System Design
2.3. Control of the Input Pulse
2.4. Design of the Cuvettes
2.5. Immobilization of the Enzyme
3. Theoretical Considerations
3.1. Flow in the Channel
3.2. Measurement in the Cuvette
4. Results and Discussion
4.1. Effect of the Cuvette on the Signal
4.2. Extraction of the Signal Produced by the Flow System
4.3. Numerical Averaging to Decrease the Noise Effects
4.4. The Quality of the Pulse Shape Reconstruction
4.5. Discussion of Signal Processing and Networking
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Molecular and Supramolecular Information Processing—From Molecular Switches to Unconventional Computing; Katz, E. (Ed.) Wiley-VCH: Weinheim, Germany, 2012.
- Szacilowski, K. Infochemistry—Information Processing at the Nanoscale; Wiley: Chichester, UK, 2012. [Google Scholar]
- De Silva, A.P. Molecular Logic-Based Computation; Royal Society of Chemistry: Cambridge, UK, 2013. [Google Scholar]
- De Silva, A.P. Molecular logic and computing. Nat. Nanotechnol. 2007, 2, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Pischel, U. Advanced molecular logic with memory function. Angew. Chem. Int. Ed. 2010, 49, 1356–1358. [Google Scholar] [CrossRef] [PubMed]
- Szacilowski, K. Digital information processing in molecular systems. Chem. Rev. 2008, 108, 3481–3548. [Google Scholar] [CrossRef] [PubMed]
- Pischel, U.; Andreasson, J.; Gust, D.; Pais, V.F. Information processing with molecules—Quo Vadis? ChemPhysChem 2013, 14, 28–46. [Google Scholar] [CrossRef] [PubMed]
- Biomolecular Information Processing—From Logic Systems to Smart Sensors and Actuators; Katz, E. (Ed.) Wiley-VCH: Weinheim, Germany, 2012.
- Katz, E. Biocomputing—Tools, aims, perspectives. Curr. Opin. Biotechnol. 2015, 34, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y. Biomolecular computing systems: Principles, progress and potential. Nat. Rev. Genet. 2012, 13, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Stefanovic, D.; Rudchenko, S. Exercises in molecular computing. Acc. Chem. Res. 2014, 47, 1845–1852. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Stefanovic, D. Chemistry at a higher level of abstraction. J. Comput. Theor. Nanosci. 2011, 8, 434–440. [Google Scholar] [CrossRef]
- Ezziane, Z. DNA computing: Applications and challenges. Nanotechnology 2006, 17, R27–R39. [Google Scholar] [CrossRef]
- Ashkenasy, G.; Dadon, Z.; Alesebi, S.; Wagner, N.; Ashkenasy, N. Building logic into peptide networks: Bottom-up and top-down. Israel J. Chem. 2011, 51, 106–117. [Google Scholar] [CrossRef]
- Katz, E.; Privman, V. Enzyme-based logic systems for information processing. Chem. Soc. Rev. 2010, 39, 1835–1857. [Google Scholar] [CrossRef] [PubMed]
- Kahan, M.; Gil, B.; Adar, R.; Shapiro, E. Towards molecular computers that operate in a biological environment. Phys. D 2008, 237, 1165–1172. [Google Scholar] [CrossRef]
- Unconventional Computing; Adamatzky, A.; De Lacy Costello, B.; Bull, L.; Stepney, S.; Teuscher, C. (Eds.) Luniver Press: Beckington, UK, 2007.
- Unconventional Computation; Calude, C.S.; Costa, J.F.; Dershowitz, N.; Freire, E.; Rozenberg, G. (Eds.) Springer: Berlin, Germany, 2009; Volume 5715.
- Claussen, J.C.; Hildebrandt, N.; Susumu, K.; Ancona, M.G.; Medintz, I.L. Complex logic functions implemented with quantum dot bionanophotonic circuits. ACS Appl. Mater. Interfaces 2014, 6, 3771–3778. [Google Scholar] [CrossRef] [PubMed]
- Privman, V. Approaches to control of noise in chemical and biochemical information and signal processing. In Molecular and Supramolecular Information Processing—From Molecular Switches to Logic Systems; Katz, E., Ed.; Wiley-VCH: Weinheim, Germany, 2012; pp. 281–303. [Google Scholar]
- Privman, V. Control of noise in chemical and biochemical information processing. Israel J. Chem. 2011, 51, 118–131. [Google Scholar] [CrossRef]
- Domanskyi, S.; Privman, V. Modeling and modifying response of biochemical processes for biocomputing and biosensing signal processing. In Advances in Unconventional Computing; Adamatzky, A., Ed.; Springer: New York, NY, USA, 2016. [Google Scholar]
- Privman, V.; Strack, G.; Solenov, D.; Pita, M.; Katz, E. Optimization of enzymatic biochemical logic for noise reduction and scalability: How many biocomputing gates can be interconnected in a circuit? J. Phys. Chem. B 2008, 112, 11777–11784. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, S.; Zavalov, O.; Halámek, J.; Privman, V.; Katz, E. Modularity of biochemical filtering for inducing sigmoid response in both inputs in an enzymatic AND gate. J. Phys. Chem. B 2013, 117, 9857–9865. [Google Scholar] [CrossRef] [PubMed]
- Domanskyi, S.; Privman, V. Design of digital response in enzyme-based bioanalytical systems for information processing applications. J. Phys. Chem. B 2012, 116, 13690–13695. [Google Scholar] [CrossRef] [PubMed]
- Halámek, J.; Zavalov, O.; Halámková, L.; Korkmaz, S.; Privman, V.; Katz, E. Enzyme-based logic analysis of biomarkers at physiological concentrations: AND gate with double-sigmoid “filter” response. J. Phys. Chem. B 2012, 116, 4457–4464. [Google Scholar] [CrossRef] [PubMed]
- Pita, M.; Privman, V.; Arugula, M.A.; Melnikov, D.; Bocharova, V.; Katz, E. Towards biochemical filter with sigmoidal response to pH changes: Buffered biocatalytic signal transduction. Phys. Chem. Chem. Phys. 2011, 13, 4507–4513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Privman, V.; Fratto, B.E.; Zavalov, O.; Halámek, J.; Katz, E. Enzymatic AND logic gate with sigmoid response induced by photochemically controlled oxidation of the output. J. Phys. Chem. B 2013, 117, 7559–7568. [Google Scholar] [CrossRef] [PubMed]
- Privman, V.; Zavalov, O.; Halámková, L.; Moseley, F.; Halámek, J.; Katz, E. Networked enzymatic logic gates with filtering: New theoretical modeling expressions and their experimental application. J. Phys. Chem. B 2013, 117, 14928–14939. [Google Scholar] [CrossRef] [PubMed]
- Privman, V.; Halámek, J.; Arugula, M.A.; Melnikov, D.; Bocharova, V.; Katz, E. Biochemical filter with sigmoidal response: Increasing the complexity of biomolecular logic. J. Phys. Chem. B 2010, 114, 14103–14109. [Google Scholar] [CrossRef] [PubMed]
- Zavalov, O.; Bocharova, V.; Halámek, J.; Halámková, L.; Korkmaz, S.; Arugula, M.A.; Chinnapareddy, S.; Katz, E.; Privman, V. Two-input enzymatic logic gates made sigmoid by modifications of the biocatalytic reaction cascades. Int. J. Unconv. Comput. 2012, 8, 347–365. [Google Scholar]
- Zavalov, O.; Bocharova, V.; Privman, V.; Katz, E. Enzyme-based logic: OR gate with double-sigmoid filter response. J. Phys. Chem. B 2012, 116, 9683–9689. [Google Scholar] [CrossRef] [PubMed]
- Halámek, J.; Zhou, J.; Halámková, L.; Bocharova, V.; Privman, V.; Wang, J.; Katz, E. Biomolecular filters for improved separation of output signals in enzyme logic systems applied to biomedical analysis. Anal. Chem. 2011, 83, 8383–8386. [Google Scholar] [CrossRef] [PubMed]
- Zavalov, O.; Domanskyi, S.; Privman, V.; Simonian, A. Design of biosensors with extended linear response and binary-type sigmoid output using multiple enzymes. In Proceedings of the Seventh International Conference on Quantum, Nano and Micro Technologies (ICQNM 2013), Barcelona, Spain, 25–31 August 2013; pp. 54–59.
- Kang, D.; Vallée-Bélisle, A.; Porchetta, A.; Plaxco, K.W.; Ricci, F. Re-engineering electrochemical biosensors to narrow or extend their useful dynamic range. Angew. Chem. Int. Ed. 2012, 51, 6717–6721. [Google Scholar] [CrossRef] [PubMed]
- Rafael, S.P.; Vallée-Bélisle, A.; Fabregas, E.; Plaxco, K.; Palleschi, G.; Ricci, F. Employing the metabolic “Branch Point Effect” to generate an all-or-none, digital-like response in enzymatic outputs and enzyme-based sensors. Anal. Chem. 2012, 84, 1076–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallée-Bélisle, A.; Ricci, F.; Plaxco, K.W. Engineering biosensors with extended, narrowed, or arbitrarily edited dynamic range. J. Am. Chem. Soc. 2012, 134, 2876–2879. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Matamoros, E.; Liu, M.; Stefanovic, D.; Stojanovic, M.N. Training a molecular automaton to play a game. Nat. Nanotechnol. 2010, 5, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Privman, V. Biomolecular computing: Learning through play. Nat. Nanotechnol. 2010, 5, 767–768. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, S.; Gerasimova, Y.V.; Guz, N.; Kolpashchikov, D.M.; Katz, E. Bridging the two worlds: A universal interface between enzymatic and DNA computing systems. Angew. Chem. Int. Ed. 2015, 54, 6562–6566. [Google Scholar] [CrossRef] [PubMed]
- Guz, N.; Fedotova, T.A.; Fratto, B.E.; Schlesinger, O.; Alfonta, L.; Kolpashchikov, D.; Katz, E. Bioelectronic interface connecting reversible logic gates based on enzyme and DNA reactions. ChemPhysChem 2016. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Wang, J.; Privman, M.; Halámek, J. Multi-analyte digital enzyme biosensors with built-in Boolean logic. Anal. Chem. 2012, 84, 5463–5469. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Katz, E. Digital biosensors with built-in logic for biomedical applications. Israel J. Chem. 2011, 51, 141–150. [Google Scholar] [CrossRef]
- Adar, R.; Benenson, Y.; Linshiz, G.; Rosner, A.; Tishby, N.; Shapiro, E. Stochastic computing with biomolecular automata. Proc. Natl. Acad. Sci. USA 2004, 101, 9960–9965. [Google Scholar] [CrossRef] [PubMed]
- Simmel, F.C. Towards biomedical applications for nucleic acid nanodevices. Nanomedicine 2007, 2, 817–830. [Google Scholar] [CrossRef] [PubMed]
- May, E.E.; Dolan, P.L.; Crozier, P.S.; Brozik, S.; Manginell, M. Towards de novo design of deoxyribozyme biosensors for GMO detection. IEEE Sens. J. 2008, 8, 1011–1019. [Google Scholar] [CrossRef]
- Von Maltzahn, G.; Harris, T.J.; Park, J.-H.; Min, D.-H.; Schmidt, A.J.; Sailor, M.J.; Bhatia, S.N. Nanoparticle self-assembly gated by logical proteolytic triggers. J. Am. Chem. Soc. 2007, 129, 6064–6065. [Google Scholar] [CrossRef] [PubMed]
- Halámková, L.; Halámek, J.; Bocharova, V.; Wolf, S.; Mulier, K.E.; Beilman, G.; Wang, J.; Katz, E. Analysis of biomarkers characteristic of porcine liver injury—From biomolecular logic gates to animal model. Analyst 2012, 137, 1768–1770. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Windmiller, J.R.; Valdés Ramírez, G.; Zhou, M.; Halámek, J.; Katz, E.; Wang, J. Enzyme-based NAND gate for rapid electrochemical screening of traumatic brain injury in serum. Anal. Chim. Acta 2011, 703, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Halámek, J.; Bocharova, V.; Wang, J.; Katz, E. Bio-logic analysis of injury biomarker patterns in human serum samples. Talanta 2011, 83, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Halámek, J.; Windmiller, J.R.; Zhou, J.; Chuang, M.-C.; Santhosh, P.; Strack, G.; Arugula, M.A.; Chinnapareddy, S.; Bocharova, V.; Wang, J.; et al. Multiplexing of injury codes for the parallel operation of enzyme logic gates. Analyst 2010, 135, 2249–2259. [Google Scholar] [CrossRef] [PubMed]
- Windmiller, J.R.; Strack, G.; Chuang, M.-C.; Halámek, J.; Santhosh, P.; Bocharova, V.; Zhou, J.; Katz, E.; Wang, J. Boolean-format biocatalytic processing of enzyme biomarkers for the diagnosis of soft tissue injury. Sens. Actuators B Chem. 2010, 150, 285–290. [Google Scholar] [CrossRef]
- Pita, M.; Zhou, J.; Manesh, K.M.; Halámek, J.; Katz, E.; Wang, J. Enzyme logic gates for assessing physiological conditions during an injury: Towards digital sensors and actuators. Sens. Actuators B Chem. 2009, 139, 631–636. [Google Scholar] [CrossRef]
- Manesh, K.M.; Halámek, J.; Pita, M.; Zhou, J.; Tam, T.K.; Santhosh, P.; Chuang, M.-C.; Windmiller, J.R.; Abidin, D.; Katz, E.; et al. Enzyme logic gates for the digital analysis of physiological level upon injury. Biosens. Bioelectron. 2009, 24, 3569–3574. [Google Scholar] [CrossRef] [PubMed]
- Adamatzky, A. Computing with waves in chemical media: Massively parallel reaction-diffusion processors. IEICE Trans. Electron. 2004, E87–C, 1748–1757. [Google Scholar]
- Adamatzky, A.; de Lacy Costello, B. On some limitations of reaction-diffusion chemical computers in relation to Voronoi diagram and its inversion. Phys. Lett. A 2003, 309, 397–406. [Google Scholar] [CrossRef]
- Toepke, M.W.; Abhyankar, V.V.; Beebe, D.J. Microfluidic logic gates and timers. Lab Chip 2007, 7, 1449–1453. [Google Scholar] [CrossRef] [PubMed]
- Scida, K.; Li, B.L.; Ellington, A.D.; Crooks, R.M. DNA detection using origami paper analytical devices. Anal. Chem. 2013, 85, 9713–9720. [Google Scholar] [CrossRef] [PubMed]
- Fratto, B.E.; Lewer, J.M.; Katz, E. Enzyme-based half-adder and half-subtractor with a modular design. ChemPhysChem 2016. [Google Scholar] [CrossRef] [PubMed]
- Fratto, B.E.; Katz, E. Controlled logic gates—Switch gate and Fredkin gate based on enzyme-biocatalyzed reactions realized in flow cells. ChemPhysChem 2016, 17, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Fratto, B.E.; Katz, E. Reversible logic gates based on enzyme-biocatalyzed reactions and realized in flow cells—Modular approach. ChemPhysChem 2015, 16, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Fratto, B.E.; Guz, N.; Katz, E. Biomolecular computing realized in parallel flow systems: Enzyme-based Double Feynman logic gate. Parallel Process. Lett. 2015, 25. [Google Scholar] [CrossRef]
- Fratto, B.E.; Roby, L.J.; Guz, N.; Katz, E. Enzyme-based logic gates switchable between OR, NXOR and NAND Boolean operations realized in a flow system. Chem. Commun. 2014, 50, 12043–12046. [Google Scholar] [CrossRef] [PubMed]
- Moseley, F.; Halámek, J.; Kramer, F.; Poghossian, A.; Schöning, M.J.; Katz, E. An enzyme-based reversible CNOT logic gate realized in a flow system. Analyst 2014, 139, 1839–1842. [Google Scholar] [CrossRef] [PubMed]
- Privman, V.; Katz, E. Can bio-inspired information processing steps be realized as synthetic biochemical processes? Phys. Status Solidi A 2015, 212, 219–228. [Google Scholar] [CrossRef]
- Katz, E.; Privman, V.; Zavalov, O. Structure of feed-forward realizations with enzymatic processes. In Proceedings of the Eighth International Conference on Quantum, Nano/Bio, and Micro Technologies, Lisbon, Portugal, 16–20 November 2014; Privman, V., Ovchinnikov, V., Eds.; ThinkMind Digital Publishing: Wilmington, NC, USA, 2014; pp. 22–27. [Google Scholar]
- MacVittie, K.; Halámek, J.; Privman, V.; Katz, E. A bioinspired associative memory system based on enzymatic cascades. Chem. Commun. 2013, 49, 6962–6964. [Google Scholar] [CrossRef] [PubMed]
- Bocharova, V.; MacVittie, K.; Chinnapareddy, S.; Halámek, J.; Privman, V.; Katz, E. Realization of associative memory in an enzymatic process: Toward biomolecular networks with learning and unlearning functionalities. J. Phys. Chem. Lett. 2012, 3, 1234–1237. [Google Scholar] [CrossRef] [PubMed]
- MacVittie, K.; Katz, E. Self-powered electrochemical memristor based on a biofuel cell—Towards memristors integrated with biocomputing systems. Chem. Commun. 2014, 50, 4816–4819. [Google Scholar] [CrossRef] [PubMed]
- MacVittie, K.; Katz, E. Electrochemical system with memimpedance properties. J. Phys. Chem. C 2013, 117, 24943–24947. [Google Scholar] [CrossRef]
- MacVittie, K.; Halámek, J.; Katz, E. Enzyme-based D-flip-flop memory system. Chem. Commun. 2012, 48, 11742–11744. [Google Scholar] [CrossRef] [PubMed]
- Pita, M.; Strack, G.; MacVittie, K.; Zhou, J.; Katz, E. Set-reset flip-flop memory based on enzyme reactions: Towards memory systems controlled by biochemical pathways. J. Phys. Chem. B 2009, 113, 16071–16076. [Google Scholar] [CrossRef] [PubMed]
- Katz, J. Introductory Fluid Machanics; Camirdge University Press: Cambridge, UK, 2010. [Google Scholar]
- Konopka, S.J.; McDuffie, B. Diffusion coefficients of ferri- and ferrocyanide ions in aqueous media, using twin-electrode thin-layer electrochemistry. Anal. Chem. 1970, 42, 1741–1746. [Google Scholar] [CrossRef]
- Doran, P.M. Bioprocess Engineering Principles; Academic Press: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Yoon, J.-Y. Introduction to Biosensors: From Electric Circuits to Immunosensors, 2nd ed.; Springer: New York, NY, USA, 2016. [Google Scholar]
- Stroock, A.D.; Whitesides, G.M. Controlling flows in microchannels with patterned surface charge and topography. Acc. Chem. Res. 2003, 36, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Pike, D.J.; Kapur, N.; Millner, P.A.; Stewart, D.I. Flow cell design for effective biosensing. Sensors 2013, 13, 58–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]










© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, A.; Fratto, B.E.; Privman, V.; Katz, E. Design of Flow Systems for Improved Networking and Reduced Noise in Biomolecular Signal Processing in Biocomputing and Biosensing Applications. Sensors 2016, 16, 1042. https://doi.org/10.3390/s16071042
Verma A, Fratto BE, Privman V, Katz E. Design of Flow Systems for Improved Networking and Reduced Noise in Biomolecular Signal Processing in Biocomputing and Biosensing Applications. Sensors. 2016; 16(7):1042. https://doi.org/10.3390/s16071042
Chicago/Turabian StyleVerma, Arjun, Brian E. Fratto, Vladimir Privman, and Evgeny Katz. 2016. "Design of Flow Systems for Improved Networking and Reduced Noise in Biomolecular Signal Processing in Biocomputing and Biosensing Applications" Sensors 16, no. 7: 1042. https://doi.org/10.3390/s16071042
APA StyleVerma, A., Fratto, B. E., Privman, V., & Katz, E. (2016). Design of Flow Systems for Improved Networking and Reduced Noise in Biomolecular Signal Processing in Biocomputing and Biosensing Applications. Sensors, 16(7), 1042. https://doi.org/10.3390/s16071042