ECG Identification For Personal Authentication Using LSTM-Based Deep Recurrent Neural Networks
Abstract
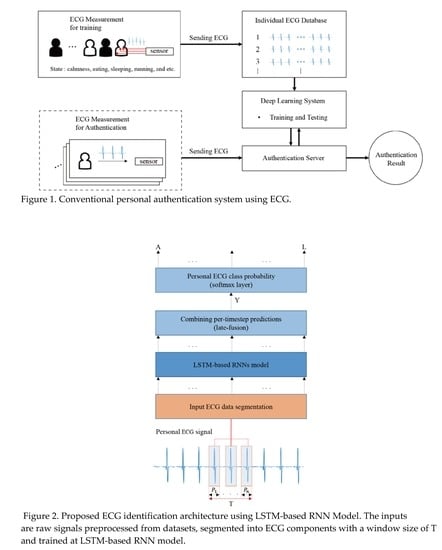
:1. Introduction
- We demonstrate the preprocessing procedures including non-feature extraction, segmentation with a fixed segmentation time period, segmentation with R-peak detection, and grouping the ECG signal of the short length. These procedures are considered for authentication time in the real-time system.
- We introduce and implement bidirectional DRNNs for ECG identification combined with the late-fusion technique. To the best of our knowledge, the proposed bidirectional DRNN model for personal authentication has not been described in the literature prior.
2. Related Work
2.1. Recurrent Neural Networks
2.2. Long Short-Term Memory (LSTM)
- Input gate () controls the input activation of new information into the memory cell.
- Output gate () controls the output flow.
- Forget gate () controls when to forget the internal state information.
- Input modulation gate () controls the main input to the memory cell.
- Internal state () controls the internal recurrence of cell.
- Hidden state () controls the information from the previous data sample within the context window:
2.3. Performance Metrics
- Precision: it calculates the number of the true person identifications (person A, B, … G) out of the positive classified classes. The overall precision (OP) is the average of the precision of each individual class (POC: the precision of each individual class):where is the true positive rate of a person classification (c =1, 2,…, c), is the false positive rate, and C is the number of classes in the dataset.
- Recall (Sensitivity): it calculates the number of persons correctly classified out of the total samples in a class. The overall recall (OR) is the average recalls for each class (RFC: recalls for each class):where is the false negative rate of a class c.
- Accuracy: it calculates the proportion of correctly predicted labels (the label is the unique name of an object) as overall predictions; an overall accuracy (OA)where, is the overall true positive rate for a classifier on all classes, is the overall true negative rate, is the overall false positive rate, and is the overall false negative rate.
- F1-score: it is the weighted average of precision and recall.where is the number of samples in a class c and is the total number of individual examples in a set of C classes.
3. Proposed Deep RNN Method and Preprocessing Procedures
3.1. Proposed Deep RNN Method
3.2. Proposed Preprocessing Procedure
3.3. Identification Procedure
3.4. Dataset and Implementation
4. Experimental Results and Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Osowski, S.; Hoai, L.; Markiewicz, T. Support Vector Machine-Based Expert System for Reliable Heartbeat Recognition. IEEE Trans. Biomed. Eng. 2004, 51, 582–589. [Google Scholar] [PubMed]
- De Chazal, P.; O’Dwyer, M.; Reilly, R. Automatic Classification of Heartbeats Using ECG Morphology and Heartbeat Interval Features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luz, E.J.D.S.; Nunes, T.M.; De Albuquerque, V.H.C.; Papa, J.P.; Menotti, D. ECG arrhythmia classification based on optimum-path forest. Expert Syst. Appl. 2013, 40, 3561–3573. [Google Scholar] [CrossRef] [Green Version]
- Inan, O.T.; Giovangrandi, L.; Kovacs, G.T. Robust neural-network-based classification of premature ventricular contractions using wavelet transform and timing interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Physionet ECG Database. Available online: www.physionet.org (accessed on 12 January 2020).
- Odinaka, I.; Lai, P.-H.; Kaplan, A.; A O’Sullivan, J.; Sirevaag, E.J.; Rohrbaugh, J.W. ECG Biometric Recognition: A Comparative Analysis. IEEE Trans. Inf. Forensics Secur. 2012, 7, 1812–1824. [Google Scholar]
- Kandala, R.N.V.P.S.; Dhuli, R.; Pławiak, P.; Naik, G.R.; Moeinzadeh, H.; Gargiulo, G.D.; Suryanarayana, G. Towards Real-Time Heartbeat Classification: Evaluation of Nonlinear Morphological Features and Voting Method. Sensors 2019, 19, 5079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, A.D.C.; Hamdy, M.M.; Badre, A.; Badee, V. Wavelet Distance Measure for Person Identification Using Electrocardiograms. IEEE Trans. Instrum. Meas. 2008, 57, 248–253. [Google Scholar] [CrossRef]
- Plataniotis, K.N.; Hatzinakos, D.; Lee, J.K.M. ECG biometric recognition without fiducial detection. In Proceedings of the 2006 Biometrics Symposium: Special Session on Research at the Biometric Consortium Conference, Baltimore, MD, USA, 19 September–21 August 2006. [Google Scholar]
- Tuncer, T.; Dogan, S.; Pławiak, P.; Acharya, U.R. Automated arrhythmia detection using novel hexadecimal local pattern and multilevel wavelet transform with ECG signals. Knowl.-Based Syst. 2019, 186, 104923. [Google Scholar] [CrossRef]
- Fratini, A.; Sansone, M.; Bifulco, P.; Cesarelli, M. Individual identification via electrocardiogram analysis. Biomed. Eng. Online 2015, 14, 78. [Google Scholar] [CrossRef] [Green Version]
- Tantawi, M.; Revett, K.; Salem, A.-B.; Tolba, M.F. ECG based biometric recognition using wavelets and RBF neural network. In Proceedings of the 7th European Computing Conference (ECC), Dubrovnik, Croatia, 25–27 June 2013; pp. 100–105. [Google Scholar]
- Zubair, M.; Kim, J.; Yoon, C. An automated ECG beat classification system using convolutional neural networks. In Proceedings of the 6th International Conference on IT Convergence and Security, Prague, Czech Republic, 26–29 September 2016; pp. 1–5. [Google Scholar]
- Pourbabaee, B.; Roshtkhari, M.J.; Khorasani, K. Deep Convolutional Neural Networks and Learning ECG Features for Screening Paroxysmal Atrial Fibrillation Patients. IEEE Trans. Syst. Man, Cybern. Syst. 2017, 99, 1–10. [Google Scholar] [CrossRef]
- Lynn, H.M.; Pan, S.B.; Kim, P. A Deep Bidirectional GRU Network Model for Biometric Electrocardiogram Classification Based on Recurrent Neural Networks. IEEE Access 2019, 7, 145395–145405. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Hammad, M.; Pławiak, P.; Wang, K.; Acharya, U.R. ResNet-Attention model for human authentication using ECG signals. Expert Syst. 2020, e12547. [Google Scholar] [CrossRef]
- Übeyli, E.D. Combining recurrent neural networks with eigenvector methods for classification of ECG beats. Digit. Signal Process. 2009, 19, 320–329. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, D.; Zeng, X. HeartID: A Multiresolution Convolutional Neural Network for ECG-Based Biometric Human Identification in Smart Health Applications. IEEE Access 2017, 5, 11805–11816. [Google Scholar] [CrossRef]
- Jasche, J.; Kitaura, F.S.; Ensslin, T.A. Digital Signal Processing in Cosmology. Digital Signal Process. 2009, 19, 320–329. [Google Scholar]
- Palangi, H.; Deng, L.; Shen, Y.; Gao, J.; He, X.; Chen, J.; Song, X.; Ward, R. Deep Sentence Embedding Using Long Short-Term Memory Networks: Analysis and Application to Information Retrieval. IEEE/ACM Trans. Audio Speech Lang. Process. 2016, 24, 694–707. [Google Scholar]
- Cho, K.; Van Merrienboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arxiv 2014, arXiv:1406.1078. [Google Scholar]
- Graves, A.; Mohamed, A.-R.; Hinton, G. Speech recognition with deep recurrent neural networks. In Proceedings of the 2013 IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP), Vancouver, BC, Canada, 26–31 May 2013; pp. 6645–6649. [Google Scholar]
- Graves, A.; Schmidhuber, J. Offline handwriting recognition with multidimensional recurrent neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 7–10 December 2009; pp. 545–552. [Google Scholar]
- Zaremba, W.; Sutskever, I.; Vinyals, O. Recurrent neural network regularization. arXiv 2014, arXiv:1409.2329. [Google Scholar]
- Salloum, R.; Kuo, C.C.J. ECG-based biometrics using recurrent neural networks. In Proceedings of the 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), New Orleans, LA, USA, 5–9 March 2017; pp. 2062–2066. [Google Scholar]
- Zhang, C.; Wang, G.; Zhao, J.; Gao, P.; Lin, J.; Yang, H. Patient-specific ECG classification based on recurrent neural networks and clustering technique. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–22 February 2017; pp. 63–67. [Google Scholar]
- Murad, A.; Pyun, J.-Y. Deep Recurrent Neural Networks for Human Activity Recognition. Sensors 2017, 17, 2556. [Google Scholar] [CrossRef] [Green Version]
- Graves, A. Supervised Sequence Labelling with Recurrent Neural Networks. Ph.D. Thesis, Technical University of Munich, Munich, Germany, 2008. [Google Scholar]
- Hochreiter, S.; Bengio, Y.; Frasconi, P.; Schmidhuber, J. Gradient flow in recurrent nets: The difficulty of learning long-term dependencies. In A Field Guide to Dynamical Recurrent Networks; IEEE Press: Piscataway, NJ, USA, 2001. [Google Scholar]
- Wu, Y.; Schuster, M.; Chen, Z.; Le, Q.V.; Norouzi, M.; Macherey, W.; Krikun, M.; Cao, Y.; Gao, Q.; Macherey, K.; et al. Google’s Neural Machine Translation System: Bridging the Gap between Human and Machine Translation. arXiv 2016, arXiv:1609.08144. [Google Scholar]
- Kittler, J.; Hatef, M.; Duin, R.P.; Matas, J. On Combining Classifiers. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 226–239. [Google Scholar] [CrossRef] [Green Version]
- Pham, V.; Bluche, T.; Kermorvant, C.; Louradour, J. Dropout Improves Recurrent Neural Networks for Handwriting Recognition. In Proceedings of the 2014 14th International Conference on Frontiers in Handwriting Recognition, Crete, Greece, 1–4 September 2014; Volume 2014, pp. 285–290. [Google Scholar]
- Yao, M. Research on Learning Evidence Improvement for kNN Based Classification Algorithm. Int. J. Database Theory Appl. 2014, 7, 103–110. [Google Scholar] [CrossRef]
- MIT-BIH Database. Available online: www.physionet.org (accessed on 12 January 2020).
- Moody, G.B.; Mark, R.G. The impact of the mit-bih arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.; Stanley, H.E. Physiobank physiotoolkit and physionet components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potdar, K.; Pardawala, T.S.; Pai, C.D. A Comparative Study of Categorical Variable Encoding Techniques for Neural Network Classifiers. Int. J. Comput. Appl. 2017, 175, 7–9. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 3, 230–236. [Google Scholar] [CrossRef]
- Yoon, H.N.; Hwang, S.H.; Choi, J.W.; Lee, Y.J.; Jeong, D.U.; Park, K.S. Slow-Wave Sleep Estimation for Healthy Subjects and OSA Patients Using R–R Intervals. IEEE J. Biomed. Health Inform. 2017, 22, 119–128. [Google Scholar] [CrossRef]
- Gacek, A.; Pedrcyz, W. ECG Signal Processing, Classification and Interpretation; Springer: London, UK, 2012. [Google Scholar]
- Azeem, T.; Vassallo, M.; Samani, N.J. Rapid Review of ECG Interpretation; Manson Publishing: Boca Raton, FL, USA, 2005. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Dar, M.N.; Akram, M.U.; Usman, A.; Khan, S.A. ECG Based Biometric Identification for Population with Normal and Cardiac Anomalies Using Hybrid HRV and DWT Features. In Proceedings of the 2015 5th International Conference on IT Convergence and Security (ICITCS), Kuala Lumpur, Malaysia, 24–27 August 2015; pp. 1–5. [Google Scholar]
- Dar, M.N.; Akram, M.U.; Shaukat, A.; Khan, M.A. ECG biometric identification for general population using multiresolution analysis of DWT based features. In Proceedings of the 2015 Second International Conference on Information Security and Cyber Forensics (Info Sec), Cape Town, South Africa, 15–17 November 2015; pp. 5–10. [Google Scholar]
- Ye, C.; Coimbra, M.T.; Kumar, B.V. Investigation of human identification using two-lead electrocardiogram (ecg) signals. In Proceedings of the 2010 Fourth IEEE International Conference on Biometrics: Theory, Applications and Systems (BTAS), Washington, DC, USA, 27–29 September 2010; pp. 1–8. [Google Scholar]
- Sidek, K.A.; Khalil, I.; Jelinek, H.F. ECG Biometric with Abnormal Cardiac Conditions in Remote Monitoring System. IEEE Trans. Syst. Man, Cybern. Syst. 2014, 44, 1498–1509. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhang, L.; Wang, H.; Tang, J. Ballistocardiogram based person identification and authentication using recurrent neural networks. In Proceedings of the 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 13–15 October 2018; pp. 1–5. [Google Scholar]
- Yildirim, Ö. A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput. Boil. Med. 2018, 96, 189–202. [Google Scholar] [CrossRef] [PubMed]











| Architectures | Layers Type |
|---|---|
| Arch 1 | Lstm-softmax |
| Arch 2 | Lstm-Lstm-softmax |
| Arch 3 | Lstm-Lstm-Lstm-softmax |
| Arch 4 | BiLstm-late-fusion-softmax |
| Arch 5 | BiLstm-BiLstm-late-fusion softmax |
| Arch 6 | BiLstm-BiLstm-BiLstm-late-fusion-softmax |
| Category | Tools |
|---|---|
| CPU | Intel i7-6700k @ 4.00 GHz |
| GPU | NVIDIA GeForce GTX 1070 @ 8GB |
| RAM | DDR4 @ 24GB |
| Operating System | Windows 10 Enterprise |
| Language | Python 3.5 |
| Library | Google Tensorflow 1.6/CUDA Toolkit 9.0/NVIDIA cuDNN v7.0 |
| Parameters | Value |
|---|---|
| Loss Function | Cross-entropy |
| Optimizer | Adam |
| Dropout | 1 |
| Learning Rate | 0.001 |
| Number of hidden units | 128 and 250 |
| Mini-batch size | 1000 and 100 |
| Type of Cell/Unit | Input Sequence Length (in Number of Beats) | Number of Hidden Layer | Overall Accuracy | Overall Precision | Overall Recall | F1 Score |
|---|---|---|---|---|---|---|
| LSTM | 2–4 | 1 | 29.7% | 24.13% | 29.68% | 0.2662 |
| LSTM | 2–4 | 2 | 98.6% | 98.73% | 98.67% | 0.9870 |
| LSTM | 2–4 | 3 | 100% | 100% | 100% | 1.0000 |
| Proposed LSTM | 2–4 | 1 | 99.93% | 99.92% | 99.96% | 0.9994 |
| Proposed LSTM | 2–4 | 2 | 99.93% | 99.92% | 99.96% | 0.9994 |
| Proposed LSTM | 2–4 | 3 | 99.93% | 99.94% | 99.93% | 0.9993 |
| Type of Cell/Unit | Input Sequence Length (in Number of Beats) | Number of Hidden Layer | Overall Accuracy | Overall Precision | Overall Recall | F1 Score |
|---|---|---|---|---|---|---|
| LSTM | 2–4 | 1 | 99.96% | 99.96% | 99.96% | 0.9996 |
| LSTM | 2–4 | 2 | 100% | 100% | 100% | 1.0000 |
| LSTM | 2–4 | 3 | 5.5% | 0.31% | 0.58% | 0.0058 |
| Proposed LSTM | 2–4 | 1 | 100% | 100% | 100% | 1.0000 |
| Proposed LSTM | 2–4 | 2 | 100% | 100% | 100% | 1.0000 |
| Proposed LSTM | 2–4 | 3 | 100% | 100% | 100% | 1.0000 |
| Type of Cell/Unit | Input Sequence Length (in Number of Beats) | Number of Hidden Layer | Overall Accuracy | Overall Precision | Overall Recall | F1 Score |
|---|---|---|---|---|---|---|
| LSTM | 0–2 | 1 | 6.28% | 7.4% | 6.21% | 0.0676 |
| LSTM | 0–2 | 2 | 38.80% | 35.66% | 38.83% | 0.3717 |
| LSTM | 0–2 | 3 | 1.87% | 0.06% | 0.18% | 0.0013 |
| Proposed LSTM | 0–2 | 1 | 81.70% | 82.83% | 81.68% | 0.9780 |
| Proposed LSTM | 0–2 | 2 | 97.78% | 97.77% | 97.77% | 0.9780 |
| Proposed LSTM | 0–2 | 3 | 98.53% | 98.53% | 98.53% | 0.9855 |
| Type of Cell/Unit | Input Sequence Length (in Number of Beats) | Number of Hidden Layer | Overall Accuracy | Overall Precision | Overall Recall | F1 Score |
|---|---|---|---|---|---|---|
| LSTM | 0–2 | 1 | 99.70% | 97.92% | 97.90% | 0.9791 |
| LSTM | 0–2 | 2 | 99.00% | 99.01% | 99.00% | 0.9900 |
| LSTM | 0–2 | 3 | 2.21% | 0.04% | 2.13% | 0.0008 |
| Proposed LSTM | 0–2 | 1 | 98.04% | 98.07% | 98.04% | 0.9806 |
| Proposed LSTM | 0–2 | 2 | 99.26% | 99.28% | 99.26% | 0.9927 |
| Proposed LSTM | 0–2 | 3 | 99.73% | 99.73% | 99.73% | 0.9973 |
| Type of Cell/Unit | Input Sequence Length (in Number of Beats) | Number of Hidden Layer | Overall Accuracy | Overall Precision | Overall Recall | F1 Score |
|---|---|---|---|---|---|---|
| LSTM | 3 | 1 | 98.65% | 98.76% | 98.85% | 0.9981 |
| LSTM | 3 | 2 | 98.17% | 98.42% | 98.56% | 0.9849 |
| LSTM | 3 | 3 | 98.55% | 98.66% | 98.86% | 0.9876 |
| LSTM | 6 | 1 | 97.00% | 97.37% | 97.49% | 0.9743 |
| LSTM | 6 | 2 | 96.85% | 97.21% | 97.61% | 0.9741 |
| LSTM | 6 | 3 | 97.92% | 98.16% | 98.44% | 0.9830 |
| LSTM | 9 | 1 | 97.50% | 97.70% | 98.07% | 0.9788 |
| LSTM | 9 | 2 | 96.50% | 96.69% | 97.11% | 0.9690 |
| LSTM | 9 | 3 | 96.49% | 96.80% | 97.22% | 0.9701 |
| Proposed LSTM | 3 | 1 | 97.79% | 98.10% | 98.22% | 0.9816 |
| Proposed LSTM | 3 | 2 | 99.37% | 99.47% | 99.52% | 0.9949 |
| Proposed LSTM | 3 | 3 | 99.20% | 99.30% | 99.42% | 0.9936 |
| Proposed LSTM | 6 | 1 | 98.71% | 98.95% | 99.06% | 0.9901 |
| Proposed LSTM | 6 | 2 | 99.57% | 99.68% | 99.59% | 0.9963 |
| Proposed LSTM | 6 | 3 | 99.71% | 99.78% | 99.72% | 0.9975 |
| Proposed LSTM | 9 | 1 | 63.80% | 67.31% | 63.49% | 0.6534 |
| Proposed LSTM | 9 | 2 | 99.10% | 99.12% | 99.31% | 0.9921 |
| Proposed LSTM | 9 | 3 | 99.80% | 99.82% | 99.83% | 0.9982 |
| Methods | Dataset | Input Sequence Length (Number of Beats) | Overall Accuracy (%) |
|---|---|---|---|
| Proposed model | MITDB | 3 | 99.73 |
| 9 | 99.80 | ||
| H. M. Lynn et al. [15] | MITDB | 3 | 97.60 |
| 9 | 98.40 | ||
| R. Salloum et al. [26] | MITDB | 3 | 98.20 |
| 9 | 100 | ||
| Q. Zhang et al. [19] | MITDB | 1 | 91.10 |
| X. Zhang [49] | MITDB | 8 | 97.80 |
| 12 | 98.9 | ||
| Ö. Yildirim [50] | MITDB | 5 | 99.39 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, B.-H.; Pyun, J.-Y. ECG Identification For Personal Authentication Using LSTM-Based Deep Recurrent Neural Networks. Sensors 2020, 20, 3069. https://doi.org/10.3390/s20113069
Kim B-H, Pyun J-Y. ECG Identification For Personal Authentication Using LSTM-Based Deep Recurrent Neural Networks. Sensors. 2020; 20(11):3069. https://doi.org/10.3390/s20113069
Chicago/Turabian StyleKim, Beom-Hun, and Jae-Young Pyun. 2020. "ECG Identification For Personal Authentication Using LSTM-Based Deep Recurrent Neural Networks" Sensors 20, no. 11: 3069. https://doi.org/10.3390/s20113069
APA StyleKim, B. -H., & Pyun, J. -Y. (2020). ECG Identification For Personal Authentication Using LSTM-Based Deep Recurrent Neural Networks. Sensors, 20(11), 3069. https://doi.org/10.3390/s20113069