Electrochemical Biosensors Based on S-Layer Proteins
Abstract
:1. Introduction
2. Bacterial S-Layer Proteins
2.1. General Features
2.2. Antifouling Properties
2.3. Electrochemical Properties
3. Basic Principles of Electrochemical Biosensors Used in Combination with S-Layer Proteins
3.1. Amperometric Biosensors
3.2. Potentiometric Biosensors
3.3. Conductometric Biosensors
3.4. Field-Effect Transistors (FETs)
3.5. Impedimetric Biosensors
4. Sensor Surface Modifications
5. Application of S-Layer Proteins in Biosensors
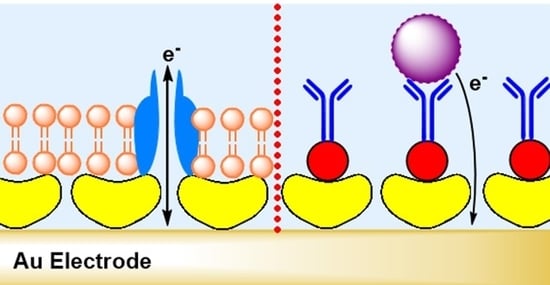
5.1. S-Layer Protein on Gold Surfaces
5.2. S-Layer Protein and Enzymes
5.3. Biosensor for Sensing Cells
5.4. S-Layer Protein and Functionalized Lipid Membrane
6. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400. [Google Scholar] [CrossRef] [PubMed]
- D’Orazio, P. Biosensors in clinical chemistry. Clin. Chim. Acta 2003, 334, 41–69. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Electrochemical biosensors for hormone analyses. Biosens. Bioelectron. 2015, 68, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, Q.; Cui, D. Recent advances in nanotechnology applied to biosensors. Sensors 2009, 9, 1033–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggins, B. Chemical sensors and biosensors. In Analytical Techniques in the Sciences; John Wiley and Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.M.; Pum, D. S-layers: Principles and applications. FEMS Microbio. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef] [PubMed]
- Pum, D.; Toca-Herrera, J.L.; Sleytr, U.B. S-Layer Protein Self-Assembly. Int. J. Mol. Sci. 2013, 14, 2484–2501. [Google Scholar] [CrossRef]
- Schuster, B.; Sleytr, U.B. Biomimetic interfaces based on S-layer proteins, lipid membranes and functional biomolecules. J. R. Soc. Interface 2014, 11, 20140232. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B. S-layer protein-based biosensors. Biosensors 2018, 8, 40. [Google Scholar] [CrossRef] [Green Version]
- Sleytr, U.B. Regular arrays of macromolecules on bacterial cell walls: Structure, chemistry, assembly, and function. Int. Rev. Cytol. 1978, 53, 1–62. [Google Scholar]
- Sleytr, U.B.; Egelseer, E.M.; Ilk, N.; Messner, P.; Schäffer, C.; Pum, D.; Schuster, B. Nanobiotechnological applications of S-layers. In Prokaryotic Cell Wall Compounds—Structure and Biochemistry; König, H., Claus, H., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.M.; Pum, D.; Horejs, C.M.; Tscheliessnig, R.; Ilk, N. Nanobiotechnology with S-Layer Proteins as Building Blocks In Progress in Molecular Biology and Translational Science, Molecular Assembly in Natural and Engineered Systems; Howorka, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Sleytr, U.B.; Messner, P. Self-assembly of crystalline bacterial cell surface layers (S-layers). In Electron Microscopy of Subcellular Dynamics; Plattner, H., Ed.; CRC Press: Boca Raton, FL, USA, 1989. [Google Scholar]
- Sleytr, U.B.; Egelseer, E.M.; Ilk, N.; Pum, D.; Schuster, B. S-layers as a basic building block in a molecular construction kit. FEBS J. 2007, 274, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.V.; Meyer, B.H. The archaeal cell envelope. Nat. Rev. Microbiol. 2011, 9, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Oliveira, T.; Belmok, A.; Vasconcellos, D.; Schuster, B.; Kyaw, C.M. Archaeal S-Layers: Overview and Current State of the Art. Front. Microbiol. 2017, 8, 2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues-Oliveira, T.; Souza, A.A.; Kruger, R.; Schuster, B.; Maria de Freitas, S.; Kyaw, C.M. Environmental factors influence the Haloferax volcanii S-layer protein structure. PLoS ONE 2019, 14, e0216863. [Google Scholar] [CrossRef]
- Mescher, M.F.; Strominger, J.L. Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 1976, 251, 2005–2014. [Google Scholar]
- Sleytr, U.B.; Thorne, K.J. Chemical characterization of the regularly arranged surface layers of Clostridium thermosaccharolyticum and Clostridium thermohydrosulfuricum. J. Bacteriol. 1976, 126, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Messner, P.; Steiner, K.; Zarschler, K.; Schäffer, C. S-layer nanoglycobiology of bacteria. Carbohydr. Res. 2008, 343, 1934–1951. [Google Scholar] [CrossRef] [Green Version]
- Eichler, J.; Maupin-Furlow, J. Post-translation modification in Archaea: Lessons from Haloferax volcanii and other haloarchaea. FEMS Microbiol. Rev. 2013, 37, 583–606. [Google Scholar] [CrossRef] [Green Version]
- Sára, M.; Sleytr, U.B. S-Layer proteins. J. Bacteriol. 2000, 182, 859–868. [Google Scholar] [CrossRef] [Green Version]
- Sára, M.; Pum, D.; Sleytr, U.B. Permeability and charge-dependent adsorption properties of the S-layer lattice from Bacillus coagulans E38–66. J. Bacteriol. 1992, 174, 3487–3493. [Google Scholar] [CrossRef] [Green Version]
- Sleytr, U.B.; Messner, P. Crystalline bacterial cell surface layers (S-layers). In Encyclopedia of Microbiology, 3rd Ed.; Schaechter, M., Ed.; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Pavkov-Keller, T.; Howorka, S.; Keller, W. The structure of bacterial S-layer proteins. In Molecular Assembly in Natural and Engineered Systems; Howorka, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Sleytr, U.B.; Beveridge, T.J. Bacterial S-layers. Trends Microbiol. 1999, 7, 253–260. [Google Scholar] [CrossRef]
- Picher, M.M.; Küpcü, S.; Huang, C.J.; Dostalek, J.; Pum, D.; Sleytr, U.B.; Ertl, P. Nanobiotechnology advanced antifouling surfaces for the continuous electrochemical monitoring of glucose in whole blood using a lab-on-a-chip. Lab Chip 2013, 13, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Györvary, E.; Pum, D. Crystallization of S-layer protein lattices on surfaces and interfaces. Prog. Organ. Coat. 2003, 47, 279–287. [Google Scholar] [CrossRef]
- Breitwieser, A.; Siedlaczek, P.; Lichtenegger, H.; Sleytr, U.B.; Pum, D. S-Layer Protein Coated Carbon Nanotubes. Coatings 2019, 9, 492. [Google Scholar] [CrossRef] [Green Version]
- Sleytr, U.B.; Egelseer, E.M.; Pum, D.; Schuster, B. S-Layers. In NanoBiotechnology: Concepts, Methods and Perspectives; Niemeyer, C.M., Mirkin, C.A., Eds.; Wiley-VCH: Weinheim, Germany, 2004. [Google Scholar]
- Fagan, R.P.; Fairweather, N.F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 2014, 12, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Pum, D.; Messner, P.; Sleytr, U.B. Role of the S layer in morphogenesis and cell division of the archaebacterium Methanocorpusculum sinense. J. Bacteriol. 1991, 173, 6865–6873. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, H.; Peters, J. Structural research on surface layers: A focus on stability, surface layer homology domains, and surface layer–cell wall interactions. J. Struct. Biol. 1998, 124, 276–302. [Google Scholar] [CrossRef]
- Zink, I.A.; Pfeifer, K.; Wimmer, E.; Sleytr, U.B.; Schuster, B.; Schleper, C. CRISPR-mediated gene silencing reveals involvement of the archaeal S-layer in cell division and virus infection. Nat. Commun. 2019, 10, 4797. [Google Scholar] [CrossRef] [Green Version]
- Györvary, E.; O’Riordan, A.; Quinn, A.J.; Redmond, G.; Pum, D.; Sleytr, U.B. Biomimetic nanostructure fabrication: Nonlithographic lateral patterning and self-assembly of functional bacterial S-layers at silicon supports. Nano Lett. 2003, 3, 315–319. [Google Scholar] [CrossRef]
- Pum, D.; Sleytr, U.B. Anisotropic crystal growth of the S-layer of Bacillus sphaericus CCM 2177 at the air/water interface. Colloids Surface A Physicochem. Eng. Asp. 1995, 102, 99–104. [Google Scholar] [CrossRef]
- Pum, D.; Sleytr, U.B. Monomolecular reassembly of a crystalline bacterial cell surface layer (S-layer) on untreated and modified silicon surfaces. Supramol. Sci. 1995, 2, 193–197. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Sára, M.; Pum, D.; Schuster, B. Crystalline bacterial cell surface layers (S-layers): A versatile self-assembly system. In Supramolecular Polymers, 2nd ed.; Ciferri, A., Ed.; Taylor and Francis Group: Milton Park, UK, 2005. [Google Scholar]
- Schuster, B.; Sleytr, U.B. Nanotechnology with S-layer proteins. In Methods in Molecular Biology; Gerrard, J.A., Domigan, L.J., Eds.; Humana Press; Springer Science+Business Media: New York, NY, USA, 2020. [Google Scholar]
- Györvary, E.; Stein, O.; Pum, D.; Sleytr, U.B. Self-assembly and recrystallization of bacterial S-layer proteins at silicon supports imaged in real time by atomic force microscopy. J. Microsc. 2003, 212, 300–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, B.; Sleytr, U.B. Biomimetic S-layer supported lipid membranes. Curr. Nanosci. 2006, 2, 143–152. [Google Scholar] [CrossRef]
- Schuster, B. Biomimetic design of nanopatterned membranes. Nanobiotechnology 2005, 1, 153–164. [Google Scholar] [CrossRef]
- Schuster, B.; Pum, D.; Sleytr, U.B. S-layer stabilized lipid membranes. Biointerphases 2008, 3, FA3–FA11. [Google Scholar] [CrossRef] [Green Version]
- Sára, M.; Manigley, C.; Wolf, G.; Sleytr, U.B. Isoporous ultrafiltration membranes from bacterial cell envelope layers. J. Membr. Sci. 1988, 36, 179–186. [Google Scholar] [CrossRef]
- Sára, M.; Sleytr, U.B. Production and characteristics of ultrafiltration membranes with uniform pores from two-dimensional arrays of proteins. J. Membr. Sci. 1987, 33, 27–49. [Google Scholar] [CrossRef]
- Pum, D.; Sára, M.; Sleytr, U.B. Two-dimensional (glyco) protein crystals as patterning elements and immobilisation matrices for the development of biosensors. In Immobilised Macromolecules: Application Potentials; Sleytr, U.B., Messner, P., Pum, D., Sára, M., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 1993. [Google Scholar]
- Sleytr, U.B.; Bayley, H.; Sára, M.; Breitwieser, A.; Küpcü, S.; Mader, C.; Weigert, S.; Unger, F.M.; Messner, P.; Jahn-Schmid, B.; et al. Applications of S-layers. FEMS Microbiol. Rev. 1997, 20, 151–175. [Google Scholar] [CrossRef] [Green Version]
- Sleytr, U.B.; Sara, M. Ultrafiltration membranes with uniform pores from crystalline bacterial cell envelope layers. Appl. Microbiol. Biot. 1986, 25, 83–90. [Google Scholar] [CrossRef]
- Sára, M.; Küpcü, S.; Weiner, C.; Weigert, S.; Sleytr, U.B. Crystalline protein layers as isoporous molecular sieves and immobilisation and affinity matrices. In Immobilised Macromolecules: Application Potentials; Sleytr, U.B., Messner, P., Pum, D., Sára, M., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 1993. [Google Scholar]
- Handrea, M.; Sahre, M.; Neubauer, A.; Sleytr, U.B.; Kautek, W. Electrochemistry of nano-scale bacterial surface protein layers on gold. Bioelectrochemistry 2003, 61, 1–8. [Google Scholar] [CrossRef]
- Sotiropoulou, S.; Mark, S.S.; Angert, E.R.; Batt, C.A. Nanoporous S-layer protein lattices. A biological ion gate with calcium selectivity. J. Phys. Chem. C 2007, 111, 13232–13237. [Google Scholar] [CrossRef]
- Zafiu, C.; Trettenhahn, G.; Pum, D.; Sleytr, U.B.; Kautek, W. Electrochemical control of adsorption dynamics of surface layer proteins on gold. Phys. Chem. Chem. Phys. 2011, 13, 3478–3483. [Google Scholar] [CrossRef] [PubMed]
- Zafiu, C.; Trettenhahn, G.; Pum, D.; Sleytr, U.B.; Kautek, W. Structural control of surface layer proteins at electrified interfaces investigated by in situ Fourier transform infrared spectroscopy. Phys. Chem. Chem. Phys. 2011, 13, 13232–13237. [Google Scholar] [CrossRef] [PubMed]
- Moudgil, A.; Kalyani, N.; Sinsinbar, G.; Das, S.; Mishra, P. S-Layer Protein for Resistive Switching and Flexible Nonvolatile Memory Device. ACS Appl. Mater. Inter. 2018, 10, 4866–4873. [Google Scholar] [CrossRef]
- Bard, A.J.; Faulkner, L.R. Electrochemical Methods, Fundamentals and Applications, 1st ed.; Wiley: Hoboken, NJ, USA, 2001. [Google Scholar]
- Juan Colás, J. Fabrication and Experimental Techniques. In Dual-Mode Electro-photonic Silicon Biosensors; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Monosik, R.; Streďanský, M.; Šturdík, E. Biosensors—Classification, characterization and new trends. Acta Chim. Slovaca 2012, 1, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Ives, D.J.G.; Janz, G.J. Reference Electrodes, Theory and Practice, 1st ed.; Academic Press: Cambridge, MA, USA, 1961. [Google Scholar]
- Narayan, R.J. Medical Biosensors For point of Care (POC) Applications; Woodhead Publishing: Sawston, UK, 2017. [Google Scholar]
- Bettazzi, F.; Marrazza, G.; Minunni, M.; Palchetti, I.; Scarano, S. Biosensors and Related Bioanalytical Tools. Compr. Anal. Chem. 2017, 77, 1–33. [Google Scholar]
- Neubauer, A.; Pum, D.; Sleytr, U.B. An amperometric glucose sensor based on isoporous crystalline protein membranes as immobilization matrix. Anal. Lett. 1993, 26, 1347–1360. [Google Scholar] [CrossRef]
- Pum, D.; Neubauer, A.; Sleytr, U.B.; Pentzien, S.; Reetz, S.; Kautek, W. Physico-chemical properties of crystalline nanoscale enzyme-protein-metal layer composites in biosensors. Ber. Bunsen. Phys. Chem. 1997, 101, 1686–1689. [Google Scholar] [CrossRef]
- Buck, R.B.; Lindner, E. Recommendations for nomenclature of ionselective electrodes. Pure Appl. Chem. 1994, 66, 2527. [Google Scholar] [CrossRef]
- Cullen, D.C.; Sethi, R.S.; Lowe, C.R. Multi-analyte miniature conductance biosensor. Anal. Chim. Acta 1990, 231, 33. [Google Scholar] [CrossRef]
- Conneely, G.; Aherne, M.; Lu, H.; Guilbault, G.G. Electrochemical immunosensors for the detection of 19-nortestosterone and methyltestosterone in bovine urine. Sens. Actuators B 2007, 121, 103–112. [Google Scholar] [CrossRef]
- Luppa, P.B.; Sokoll, L.J.; Chan, D.W. Immunosensors—Principles and applications to clinical chemistry. Clin. Chim. Acta 2001, 314, 1–26. [Google Scholar] [CrossRef]
- Willner, I.; Katz, E. Bioelectronics: From Theory to Applications; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005. [Google Scholar]
- Damiati, S.; Schrems, A.; Sinner, E.K.; Sleytr, U.B.; Schuster, B. Probing peptide and protein insertion in a biomimetic S-layer supported lipid membrane platform. Int. J. Mol. Sci. 2015, 16, 2824–2838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damiati, S.; Zayni, S.; Schrems, A.; Kiene, E.; Sleytr, U.B.; Chopineau, J.; Schuster, B.; Sinner, E.K. Inspired and stabilized by nature: Ribosomal synthesis of the human voltage gated ion channel (VDAC) into 2D-protein-tethered lipid interfaces. Biomater. Sci. 2015, 3, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Gufler, P.C.; Pum, D.; Sleytr, U.B.; Schuster, B. Highly robust lipid membranes on crystalline S-layer supports investigated by electrochemical impedance spectroscopy. Biochim. Biophys. Acta 2004, 1661, 154–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damiati, S.; Küpcü, S.; Peacock, M.; Eilenberger, C.; Zamzami, M.; Qadri, I.; Choudhry, H.; Sleytr, U.B.; Schuster, B. Acoustic and hybrid 3D-printed electrochemical biosensors for the real-time immunodetection of liver cancer cells (HepG2). Biosens. Bioelectron. 2017, 94, 500–506. [Google Scholar] [CrossRef]
- Damiati, S.; Peacock, M.; Sleytr, U.; Schuster, B. Bioinspired Diagnostic Sensor Based on Functional Nanostructures of S-Proteins to Target the Folate Receptors in Breast Cancer Cells. Sens. Actuators B 2018, 267, 224–230. [Google Scholar] [CrossRef]
- Sotiropoulou, S.; Gavalas, V.; Vamvakaki, V.; Chaniotakis, N.A. Novel carbon materials in biosensor systems. Biosens. Bioelectron. 2003, 18, 211–215. [Google Scholar] [CrossRef]
- Kuznetsov, B.A.; Shumakovich, G.P.; Koroleva, O.V.; Yaropolov, A.I. On applicability of laccase as label in the mediated and mediatorless electroimmunoassay: Effect of distance on the direct electron transfer between laccase and electrode. Biosens. Bioelectron. 2001, 16, 73–84. [Google Scholar] [CrossRef]
- Reimhult, E.; Höök, F. Design of Surface Modifications for Nanoscale Sensor Applications. Sensors 2015, 15, 1635. [Google Scholar] [CrossRef] [Green Version]
- Guidelli, R.; Becucci, L. Electrochemistry of Biomimetic Membranes. In Applications of Electrochemistry and Nanotechnology in Biology and Medicine II. Modern Aspects of Electrochemistry; Eliaz, N., Ed.; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Damiati, S. Can We Rebuild the Cell Membrane. In Biological, Physical and Technical Basics of Cell Engineering; Artmann, G., Artmann, A., Zhubanova, A., Digel, I., Eds.; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Cremer, P.S.; Boxer, S.G. Formation and spreading of lipid bilayers on planar glass supports. J. Phys. Chem. B 1999, 103, 2554–2559. [Google Scholar] [CrossRef]
- Hardy, G.J.; Nayak, R.; Zauscher, S. Model cell membranes: Techniques to form complex biomimetic supported lipid bilayers via vesicle fusion. Curr. Opin. Colloid Interface Sci. 2013, 18, 448–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, N.J.; Frank, C.W.; Kasemo, B.; Hook, F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat. Protoc. 2010, 5, 1096–1106. [Google Scholar] [CrossRef] [PubMed]
- Tiefenauer, L.; Demarche, S. Challenges in the development of functional assays of membrane proteins. Materials 2012, 5, 2205. [Google Scholar] [CrossRef] [Green Version]
- Jackson, A.; Boutell, J.; Cooley, N.; He, M. Cell-free protein synthesis for proteomics. Brief Funct. Genom. Proteom. 2004, 2, 308–319. [Google Scholar] [CrossRef] [Green Version]
- Endo, Y.; Sawasaki, T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 2006, 17, 373–380. [Google Scholar] [CrossRef]
- Damiati, S.; Mhanna, R.; Kodzius, R.; Ehmoser, E.K. Cell-Free Approaches in Synthetic Biology Utilizing Microfluidics. Genes 2018, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Kalia, J.; Raines, R.T. Advances in Bioconjugation. Curr. Org. Chem. 2010, 14, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Eggins, B.R. Biosensors: An Introduction; Springer-Verlag: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Fang, Y.; Ramasamy, R. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [Green Version]
- Perez, J.A.C.; Sosa-Hernandez, J.E.; Hussain, S.M.; Bilal, M.; Parra-Saldivar, R.; Iqbal, H.M.N. Bioinspired biomaterials and enzyme-based biosensors for point-of-care applications with reference to cancer and bio-imaging. Biocat. Agricult. Biotech. 2019, 17, 168–176. [Google Scholar] [CrossRef]
- Conroy, D.J.R.; Millner, P.A.; Stewart, D.I.; Pollmann, K. Biosensing for the environment and defence: Aqueous uranyl detection using bacterial surface layer proteins. Sensors 2010, 10, 4739. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, A.; Hödl, C.; Pum, D.; Sleytr, U.B. A multistep enzyme sensor for sucrose based on S-layer microparticles as immobilization matrix. Anal. Lett. 1994, 27, 849–865. [Google Scholar] [CrossRef]
- Guimarães, J.A.; Ferraz, H.C.; Alves, T.L.M. Langmuir-Blodgett films of cholesterol oxidase and S-layer proteins onto screen-printed electrodes. Appl. Surf. Sci. 2014, 298, 68–74. [Google Scholar] [CrossRef]
- Park, T.J.; Lee, S.J.; Park, J.P.; Yang, M.H.; Choi, J.H.; Lee, S.Y. Characterization of a bacterial self-assembly surface layer protein and its application as an electrical nanobiosensor. J. Nanosci. Nanotechno. 2011, 11, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Sára, M.; Küpcü, S.; Weiner, C.; Weigert, S.; Sleytr, U.B. S-layers as immobilization and affinity matrices. In Advances in Bacterial Paracrystalline Surface Layers; Beveridge, T.J., Koval, S.F., Eds.; Plenum Press: New York, NY, USA, 1993. [Google Scholar]
- Neubauer, A.; Pum, D.; Sleytr, U.B. Fibre-optic glucose biosensor using enzyme membranes with 2-D crystalline structure. Biosens. Bioelectron. 1996, 11, 317–325. [Google Scholar] [CrossRef]
- Neubauer, A.; Pentzien, S.; Reetz, S.; Kautek, W.; Pum, D.; Sleytr, U.B. Pulsed-laser metal contacting of biosensors on the basis of crystalline enzyme-protein layer composites. Sens. Actuators B Chem. 1997, 40, 231–236. [Google Scholar] [CrossRef]
- Schuster, B.; Sleytr, U.B. Relevance of glycosylation of S-layer proteins for cell surface properties. Acta Biomater. 2015, 19, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Hao, J.; Yang, S.; Sun, Y.; Wang, Y.; Zhang, W.; Mao, H.; Song, X.-M. Layer-by-layer self-assembly film of PEI-reduced graphene oxide composites and cholesterol oxidase for ultrasensitive cholesterol biosensing. Sens. Actuators B Chem. 2019, 298, 126856. [Google Scholar] [CrossRef]
- Ferraz, H.C.; Guimarães, J.A.; Alves, T.L.M.; Constantino, C.J.L. Monomolecular films of cholesterol oxidase and S-Layer proteins. Appl. Surf. Sci. 2011, 257, 6535–6539. [Google Scholar] [CrossRef]
- El-Said, W.A.; Yea, C.H.; Kim, H.; Oh, B.K.; Choi, J.W. Cell-based chip for the detection of anticancer effect on HeLa cells using cyclic voltammetry. Biosens. Bioelectron. 2009, 24, 1259–1265. [Google Scholar] [CrossRef]
- Yeon, J.H.; Park, J.-K. Cytotoxicity test based on electrochemical impedance measurement of HepG2 cultured in microfabricated cell chip. Anal. Biochem. 2005, 341, 308–315. [Google Scholar] [CrossRef]
- Zhu, X.; Feng, C.; Ye, Z.; Chen, Y.; Li, G. Fabrication of magneto-controlled moveable architecture to develop reusable electrochemical biosensors. Sci. Rep. 2015, 4, 4169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damiati, S.; Peacock, M.; Leonhardt, S.; Damiati, L.; Baghdadi, M.A.; Becker, H.; Kodzius, R.; Schuster, B. Embedded Disposable Functionalized Electrochemical Biosensor with a 3D-Printed Flow-Cell for Detection of Hepatic Oval Cells. Genes 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Lu, J.; Chen, Z.; Yu, Y.; Mo, M. A repeatable assembling and disassembling electrochemical aptamer cytosensor for ultrasensitive and highly selective detection of human liver cancer cells. Anal. Chim. Acta 2015, 885, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Liu, P.; Petrenko, V.A.; Liu, A. A Label-Free Electrochemical Impedance Cytosensor Based on Specific Peptide-Fused Phage Selected from Landscape Phage Library. Sci. Rep. 2016, 6, 22199. [Google Scholar] [CrossRef] [Green Version]
- Baniukevic, J.; Kirlyte, J.; Ramanavicius, A.; Ramanaviciene, A. Application of oriented and random antibody immobilization methods in immunosensor design. Sens. Actuators B Chem. 2013, 189, 217–223. [Google Scholar] [CrossRef]
- Deng, X.; Chen, M.; Fu, Q.; Smeets, N.; Fu, X.; Zhang, Z.; Filipe, C.; Hoare, T. A Highly Sensitive Immunosorbent Assay Based on Biotinylated Graphene Oxide and the Quartz Crystal Microbalance. ACS Appl. Mater. Interfaces 2016, 8, 1893–1902. [Google Scholar] [CrossRef]
- Bae, Y.; Oh, M.; Lee, W.; Lee, W.; Choi, J. Study on orientation of immunoglobulin G on protein G layer. Biosens. Bioelectron. 2005, 21, 103–110. [Google Scholar] [CrossRef]
- Kausaite-Minkstimiene, A.; Ramanaviciene, A.; Kirlyte, J.; Ramanavicius, A. Comparative Study of Random and Oriented Antibody Immobilization Techniques on the Binding Capacity of Immunosensor. Anal. Chem. 2010, 82, 6401–6408. [Google Scholar] [CrossRef]
- Xiao, Y.; Lin, M.; Jiang, X.; Ye, J.; Guo, T.; Shi, Y.; Bian, X. The Recent Advances on Liver Cancer Stem Cells: Biomarkers, Separation, and Therapy. Anal. Cell Pathol. 2017, 2017, 5108653. [Google Scholar] [CrossRef] [Green Version]
- Eliasson, M.; Olsson, A.; Palmcrantz, E.; Wiberg, K.; Inganas, M.; Guss, B.; Lindberg, M.; Uhlen, M. Chimeric IgG-binding receptors engineered from staphylococcal protein A and streptococcal protein. J. Biol. Chem. 1988, 263, 4323–4327. [Google Scholar]
- Völlenkle, C.; Weigert, S.; Ilk, N.; Egelseer, E.; Weber, V.; Loth, F.; Falkenhagen, D.; Sleytr, U.B.; Sara, M. Construction of a Functional S-Layer Fusion Protein Comprising an Immunoglobulin G-Binding Domain for Development of Specific Adsorbents for Extracorporeal Blood Purification. Appl. Environ. Microbiol. 2004, 70, 1514–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rosa, M. Archaeal lipids: Structural features and supramolecular organization. Thin Solid Films 1996, 284–285, 13–17. [Google Scholar] [CrossRef]
- Hanford, M.J.; Peeples, T.L. Archaeal tetraether lipids: Unique structures and applications. Appl. Biochem. Biotechnol. Enzym. Eng. Biotechnol. 2002, 97, 45–62. [Google Scholar] [CrossRef]
- Eme, L.; Spang, A.; Lombard, J.; Stairs, C.W.; Ettema, T.J.G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2017, 15, 711–723. [Google Scholar] [CrossRef]
- Engelhardt, H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 2007, 160, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, H. Mechanism of osmoprotection by archaeal S-layers: A theoretical study. J. Struct. Biol. 2007, 160, 190–199. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.; Sleytr, U.B. Composite S-layer lipid structures. J. Struct. Biol. 2009, 168, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.; Pum, D.; Sleytr, U.B. Voltage clamp studies on S-layer-supported tetraether lipid membranes. Biochim. Biophys. Acta 1998, 1369, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Abbasi, F.; Leitch, J.J.; Faragher, R.J.; Schwan, A.L.; Lipkowski, J. Mechanisms of alamethicin ion channel inhibition by amiloride in zwitterionic tethered lipid bilayers. J. Electroanal. Chem. 2019, 848, 113281. [Google Scholar] [CrossRef]
- Schrems, A.; Larisch, V.D.; Stanetty, C.; Dutter, K.; Damiati, S.; Sleytr, U.B.; Schuster, B. Liposome fusion on proteinaceous S-layer lattices triggered via β-diketone ligand-europium(iii) complex formation. Soft Matter 2011, 7, 5514–5518. [Google Scholar] [CrossRef]
- Colombini, M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 2004, 256–257, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Colombini, M. VDAC structure, selectivity, and dynamics. Biochim. Biophys. Acta 2012, 1818, 1457–1465. [Google Scholar] [CrossRef] [Green Version]
- Colombini, M. The VDAC channel: Molecular basis for selectivity. Biochim. Biophys. Acta 2016, 1863, 2498–2502. [Google Scholar] [CrossRef] [PubMed]
- Colombini, M.; Blachly-Dyson, E.; Forte, M. VDAC, a channel in the outer mitochondrial membrane. Ion Channels 1996, 4, 169–202. [Google Scholar]
- Komarov, A.G.; Deng, D.; Craigen, W.J.; Colombini, M. New insights into the mechanism of permeation through large channels. Biophys. J. 2005, 89, 3950–3959. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.C.; Xu, X.; Colombini, M. The role of pyridine dinucleotides in regulating the permeability of the mitochondrial outer membrane. J. Biol. Chem. 1996, 271, 26724–26731. [Google Scholar] [CrossRef] [Green Version]
- Rostovtseva, T.K.; Komarov, A.; Bezrukov, S.M.; Colombini, M. Dynamics of nucleotides in VDAC channels: Structure-specific noise generation. Biophys. J. 2002, 82, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Zizi, M.; Forte, M.; Blachly-Dyson, E.; Colombini, M. NADH regulates the gating of VDAC, the mitochondrial outer membrane channel. J. Biol. Chem. 1994, 269, 1614–1616. [Google Scholar]
- Knoll, W.; Naumann, R.; Friedrich, M.; Robertson, J.W.F.; Lösche, M.; Heinrich, F.; McGillivray, D.J.; Schuster, B.; Gufler, P.C.; Pum, D.; et al. Solid supported lipid membranes: New concepts for the biomimetic functionalization of solid surfaces. Biointerphases 2008, 3, FA125–FA135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, W.; Köper, I.; Naumann, R.; Sinner, E.K. Tethered bimolecular lipid membranes-A novel model membrane platform. Electrochim. Acta 2008, 53, 6680–6689. [Google Scholar] [CrossRef]
- Sinner, E.K.; Knoll, W. Functional tethered membranes. Curr. Opin. Chem. Biol. 2001, 5, 705–711. [Google Scholar] [CrossRef]
- Sinner, E.K.; Ritz, S.; Naumann, R.; Schiller, S.; Knoll, W. Self-assembled tethered bimolecular lipid membranes. Adv. Clin. Chem. 2009, 49, 159–179. [Google Scholar] [PubMed]
- Zaba, C.; Ritz, S.; Tan, C.-W.D.; Zayni, S.; Müller, M.; Reuning, U.; Sinner, E.K. Functional Cell Adhesion Receptors (Integrins) in Polymeric Architectures. ChemBioChem 2015, 16, 1740–1743. [Google Scholar] [CrossRef] [PubMed]
- Zapf, T.; Tan, C.-W.D.; Reinelt, T.; Huber, C.; Shaohua, D.; Geifman-Shochat, S.; Paulsen, H.; Sinner, E.K. Synthesis and Functional Reconstitution of Light-Harvesting Complex II into Polymeric Membrane Architectures. Angew. Chem. Int. Edit. 2015, 54, 14664–14668. [Google Scholar] [CrossRef]
- Czernohlavek, C.; Schuster, B. Formation and characteristics of mixed lipid/polymer membranes on a crystalline surface-layer protein lattice. Biointerphases 2020, 15, 011002. [Google Scholar] [CrossRef] [Green Version]
- Czernohlavek, C.; Schuster, B. Formation of planar hybrid lipid/polymer membranes anchored to an S-layer protein lattice by vesicle binding and rupture. Soft Mater. 2020. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.; Sleytr, U.B. Tailor-made crystalline structures of truncated S-layer proteins on heteropolysaccharides. Soft Matter 2009, 5, 334–341. [Google Scholar] [CrossRef]
- Jackman, J.A.; Knoll, W.; Cho, N.-J. Biotechnology applications of tethered lipid bilayer membranes. Materials 2012, 5, 2637. [Google Scholar] [CrossRef] [Green Version]
- Trojanowicz, M. Miniaturized biochemical sensing devices based on planar lipid membranes. Fresenius J. Anal. Chem. 2001, 372, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.D.; McAllister, G.; Feighner, S.D.; Liu, Q.; Nargund, R.P.; Van der Ploeg, L.H.T.; Patchett, A.A. Orphan G-protein-coupled receptors and natural ligand discovery. Trends Pharmacol. Sci. 2001, 22, 132–140. [Google Scholar] [CrossRef]
- Howard, A.D.; Wang, R.; Pong, S.S.; Mellin, T.N.; Strack, A.; Guan, X.M.; Zeng, Z.; Williams Jr, D.L.; Feighner, S.D.; Nunes, C.N.; et al. Identification of receptors for neuromedin U and its role in feeding. Nature 2000, 406, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.B. Targeting cardiac potassium channels for state-of-the-art drug discovery. Expert Opin. Drug Dis. 2015, 10, 157–169. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, Y.; Cui, H.; Du, X. Bio-inspired sensing and actuating materials. J. Mater. Chem. C 2019, 7, 6493–6511. [Google Scholar] [CrossRef]
- Liao, Z.; Wang, J.; Zhang, P.; Zhang, Y.; Miao, Y.; Gao, S.; Deng, Y.; Geng, L. Recent advances in microfluidic chip integrated electronic biosensors for multiplexed detection. Biosens. Bioelectron. 2018, 121, 272–280. [Google Scholar] [CrossRef]
- Naranjo-Hernández, D.; Reina-Tosina, J.; Min, M. Fundamentals, recent advances, and future challenges in bioimpedance devices for healthcare applications. J. Sensors 2019, 9210258. [Google Scholar] [CrossRef] [Green Version]
- Primiceri, E.; Chiriacò, M.S.; Notarangelo, F.M.; Crocamo, A.; Ardissino, D.; Cereda, M.; Bramanti, A.P.; Bianchessi, M.A.; Giannelli, G.; Maruccio, G. Key enabling technologies for point-of-care diagnostics. Sensors 2018, 18, 3607. [Google Scholar] [CrossRef] [Green Version]
- Jurado-Sánchez, B. Microscale and nanoscale biosensors. Biosensors 2018, 8, 66. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef]







| Molecular weight of S-layer protein subunits: 40–200,000 Da | [7,23,24,25,26,27,28] |
| Reactive groups (e.g., carboxyl- and amino-residues) occur on each protomer in identical position and orientation | [7,23,24] |
| Two-dimensional (glyco)protein crystal composed of identical subunits | [7,25] |
| Oblique (p2), square (p4) or hexagonal (p6) space group symmetry | [7,16,26,27] |
| Center-to-center spacing of unit cells (= morphological units) of crystalline lattice: 3.5–35 nm | [7,27] |
| Layer thickness: 5–10 nm | [7,8] |
| High porosity (30%-70%) with pores of identical size (2–8 nm), morphology, and physicochemical properties | [7,24] |
| Topography: Inner surface smooth, outer surface more corrugated | [7,8,26] |
| Anisotropic charge distribution between outer and inner face: Outer face charge neutral due to an equal number of carboxyl- and amino groups. Inner face net negatively charged due to an excess of carboxyl groups | [7,8,24] |
| Antifouling, non-sticky outer surface | [7,11,28] |
| Self-assembly capability in aqueous media, on the air/water interface, on lipid films, and on solid surfaces like metals (gold, silver, platinum, stainless steel), glass, silicon, silicon oxide and nitride, mica, polymers (e.g., polystyrene, polyester, cellulose, polydimethylsiloxane (PDMS), indium tin oxide (ITO), highly oriented pyrolytic graphite (HOPG), and carbon nanotubes | [7,9,28,29,30] |
| Electrode Architecture | Immobilization Method | Biorecognition Element | Detected Species | Electro-Chemical Method | Tested Detection Range | Linear Range | Stability | Remark | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Au-SAM- SLP | Maleimide-Cys; biotin-avidin | SLP | UO22+ | EIS | 10−5–10−12 M | 10−5–10−8 M | N.D. | LOD 10−12 M | [91] |
| Au/SLP-GOx | Chemical (EDC) | GOx + FCN | glucose | Amperometric | 0.5–50 mM | 0.5–50 mM | 2.2 h | blood, HSA, plasma | [28] |
| Au/Pt-GOx-SUM | Chemical (EDC) | GOx | glucose | Amperometric | 2–20 mM | Up to 12 mM | 48 h | Response 10–30 s | [62] |
| Au-AlcOx-SUM | Chemical (EDC) | alcohol oxidase | ethanol | Amperometric | - | Up to 7 mM | N.D. | Signal: 2.5 µA cm−2mM−1 | [63] |
| Au-XanOx-SUM | Chemical (EDC) | xanthine oxidase | xanthine | Amperometric | - | Up to 0.6 mM | N.D. | Signal: 30 µA cm−2mM−1 | [64] |
| Au-Maltase/ GOx-SUM | Chemical (EDC) | maltase + GOx | maltose | Amperometric | - | Up to 1.5 mM | N.D. | Signal: 1.5 µA cm−2mM−1 | [64] |
| Au-Inv/Mut /GOx-SUM | Chemical (EDC) | invertase + mutarotase + GOx | sucrose | Amperometric | 1–35 mM | Up to 30 mM | 36 h | Response 300 s | [92] |
| C-ChOx/ SLP | Mixed multi-layers | ChOx | cholesterol | CV | 3.1 mM | N.D. | N.D. | Langmuir/ Blodgett | [93] |
| Au-SLP/ZZ-anti-Ab | ZZ-domain + anti-CD133 | anti-CD133 antibody | Liver cancer cells (HepG2) | CV | 1 × 105–6 × 106 cells | Up to 6x106 cells | N.D. | S-layer fusion protein | [72] |
| Au-SLP-folate | Chemical (EDC) | folate | Breast cancer cells (MFC-7) | SWV | 1 × 104–5 × 105 cells | N.D. | N.D. | LOD 1 × 105 cells/mL | [73] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damiati, S.; Schuster, B. Electrochemical Biosensors Based on S-Layer Proteins. Sensors 2020, 20, 1721. https://doi.org/10.3390/s20061721
Damiati S, Schuster B. Electrochemical Biosensors Based on S-Layer Proteins. Sensors. 2020; 20(6):1721. https://doi.org/10.3390/s20061721
Chicago/Turabian StyleDamiati, Samar, and Bernhard Schuster. 2020. "Electrochemical Biosensors Based on S-Layer Proteins" Sensors 20, no. 6: 1721. https://doi.org/10.3390/s20061721
APA StyleDamiati, S., & Schuster, B. (2020). Electrochemical Biosensors Based on S-Layer Proteins. Sensors, 20(6), 1721. https://doi.org/10.3390/s20061721