First Contact to Odors: Our Current Knowledge about Odorant Receptor
Abstract
:1. Introduction
2. Odorant Receptors in Mammals
2.1. Anatomy of the olfactory system
2.2. Olfactory signal transduction
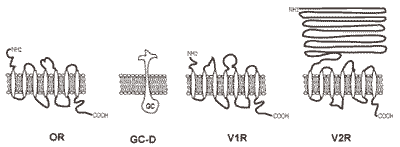
2.3. Odorant receptors
2.4. Expression of odorant receptors in the olfactory epithelium
2.5. Heterologous expression of odorant receptors
2.6. Pheromone receptors
2.7. Atypical odorant receptors
2.8. Combinational odorant receptor coding for odors
3. Odorant receptors in insects
3.1. Odorant receptors and olfactory system in Drosophila
3.2. Olfactory organs in Drosophila
3.3. Olfactory signal transduction in Drosophila (Figure 6)
3.4. Expression of odorant receptor genes in Drosophila
3.5. Function of odorant receptors in Drosophila
3.6. Carbon dioxide receptors in Drosophila
4. Conclusions
Acknowledgments
References
- Radhika, V.; Proikas-Cezanne, T.; Jayaraman, M.; Onesime, D.; Ha, J.H.; Dhanasekaran, D.N. Chemical sensing of DNT by engineered olfactory yeast strain. Nat. Chem. Biol. 2007, 3, 325–330. [Google Scholar]
- Buck, L.B. The search for odorant receptors. Cell 2004, 116, 117–119. [Google Scholar]
- Farbman, A.I. Development and Plasticity in Cell Biology of Olfaction; Barlow, P.W., Bray, D., Green, P.B., Slack, J.M.W., Eds.; Cambridge University Press: Cambridge, USA, 1992; pp. 167–206. [Google Scholar]
- Labarca, P.; Bacigalupo, J. Ion channels from chemosensory olfactory neurons. J. Bioenerg. Biomem. 1988, 20, 551–569. [Google Scholar]
- Getchell, T.V. Functional properties of vertebrate olfactory receptor neurons. Physiol. Rev. 1986, 66, 772–818. [Google Scholar]
- Lowe, G.; Gold, G.H. Contribution of the ciliary cyclic nucleotide-gated conductance to olfactory transduction in the salamander. J. Physiol. 1993, 462, 175–196. [Google Scholar]
- Getchell, T.V.; Margolis, F.L.; Getchell, M.L. Perireceptor and receptor events in vertebrate olfaction. Prog. Neurobiol. 1985, 23, 317–345. [Google Scholar]
- Okano, T.M. Secreation and electrogenesis of the supporting cell in the olfactory epithelium. J. Physiol. (London) 1974, 242, 353–370. [Google Scholar]
- Graziadei, P.P.C.; Monti-Graziadei, G.A. Neurogenesis and neuron regeneration in the olfactory system of mammals. J. Neurocytol. 1979, 8, 1–18. [Google Scholar]
- Moulton, D.G.; Beidler, L.M. Structure and function in the peripheral olfactory system. Physiol. Rev. 1967, 47, 1–52. [Google Scholar]
- Caggiano, M.; Kauer, J.S.; Hunter, D.D. Globose basal cells are neuronal progenitors in the olfactory epithelium: A lineage analysis using a replication-incompetent retrovirus. Neuron 1994, 13, 339–352. [Google Scholar]
- Rhein, L.D.; Cagan, R.H. Biochemical studies of olfaction: isolation, characterization and odorant binding activity of cilia from rainbow trout olfactory rosettes. Proc. Natl. Acad. Sci. USA 1980, 77, 4412–4416. [Google Scholar]
- Buck, L.B. Information coding in the vertebrate olfactory system. Ann. Rev. Neurosci. 1996, 19, 517–544. [Google Scholar]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713–723. [Google Scholar]
- Dwyer, N.D.; Troemel, E.R.; Sengupta, P.; Bargmann, C.I. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell 1998, 93, 455–466. [Google Scholar]
- Pace, U.; Hanski, E.; Salomon, Y.; Lancet, D. Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature 1985, 316, 255–258. [Google Scholar]
- Sklar, P.B.; Anholt, R.R.H.; Snyder, S.H. The odorant-sensitive adenylate cyclase of olfactory receptor neurons. J. Biol. Chem. 1986, 261, 15538–15543. [Google Scholar]
- Ronnett, G.V.; Cho, H.; Hester, L.D.; Wood, S.F.; Snyder, S.H. Odorants differentially enhance phosphoinositide turnover and adenylyl cyclase in olfactory receptor neuronal cultures. J. Neurosci. 1993, 13, 1751–1758. [Google Scholar]
- Nakamura, T.; Gold, G.H. A cyclic-nucleotide gated conductance in olfactory receptor cilia. Nature 1987, 325, 442–444. [Google Scholar]
- Firestein, S.; Werblin, F.S. Odor-induced membrane currents in vertebrate olfactory receptor neurons. Science 1989, 244, 79–82. [Google Scholar]
- Getchell, T.V.; Shepherd, G.M. Adaptive properties of olfactory receptor analysed with odour pulses of varying durations. J. Physiol. 1978, 282, 541–560. [Google Scholar]
- Ottoson, D. Analysis of the electrical activity of the olfactory epithelium. Acta. Physiol. Scan 1956, 122, 1–83. [Google Scholar]
- Pelosi, P.; Baldaccini, N.E.; Pisanelli, A.M. Identification of a specific olfactory receptor for 2-isobutyl-3-methoxypyrazine. Biochem. J. 1982, 201, 245–248. [Google Scholar]
- Pevsner, J.; Trifiletti, R.R.; Strittmatter, S.S.; Snyder, S.H. Isolation and characterization of an olfactory receptor protein for odorant pyrazines. Proc. Natl. Acad. Sci. USA 1985, 82, 3050–3054. [Google Scholar]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar]
- Mombaerts, P. Molecular biology of odorant receptors in vertebrates. Annu. Rev. Neurosci. 1999, 22, 487–509. [Google Scholar]
- Glusman, G.; Yanai, I.; Rubin, I.; Lancet, D. The complete human olfactory subgenome. Genome. Res. 2001, 11, 685–702. [Google Scholar]
- Raming, K.; Konzelmann, S.; Breer, H. Identification of a novel G-protein coupled receptor expressed in distinct brain regions and a defined olfactory zone. Receptors Channels 1998, 6, 141–151. [Google Scholar]
- Feingold, E.A.; Penny, L.A.; Nienhuis, A.W.; Forget, B.G. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics 1999, 61, 15–23. [Google Scholar]
- Mombaerts, P. Odorant receptor genes in humans. Curr. Opin. Genet. Dev. 1999, 9, 315–320. [Google Scholar]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; Gocayne, J.D.; Amanatides, P.; Ballew, R.M.; Huson, D.H.; Wortman, J.R.; Zhang, Q.; Kodira, C.D.; Zheng, X.H.; Chen, L.; Skupski, M.; Subramanian, G.; Thomas, P.D.; Zhang, J.; Miklos, G.L.G.; Nelson, C.; Broder, S.; Clark, A.G.; Nadeau, J.; McKusick, V.A.; Zinder, N.; Levine, A.J.; Roberts, R.J.; Simon, M.; Slayman, C.; Hunkapiller, M.; Bolanos, R.; Delcher, A.; Dew, I.; Fasulo, D.; Flanigan, M.; Florea, L.; Halpern, A.; Hannenhalli, S.; Kravitz, S.; Levy, S.; Mobarry, C.; Reinert, K.; Remington, K.; Abu-Threideh, J.; Beasley, E.; Biddick, K.; Bonazzi, V.; Brandon, R.; Cargill, M.; Chandramouliswaran, I.; Charlab, R.; Chaturvedi, K.; Deng, Z.; Di Francesco, V.; Dunn, P.; Eilbeck, K.; Evangelista, C.; Gabrielian, A.E.; Gan, W.; Ge, W.; Gong, F.; Gu, Z.; Guan, P.; Heiman, T.J.; Higgins, M.E.; Ji, R.; Ke, Z.; Ketchum, K.A.; Lai, Z.; Lei, Y.; Li, Z.; Li, J.; Liang, Y.; Lin, X.; Lu, F.; Merkulov, G.V.; Milshina, N.; Moore, H.M.; Naik, A.K.; Narayan, V.A.; Neelam, B.; Nusskern, K.; Rusch, D.B.; Salzberg, S.; Shao, W.; Shue, B.; Sun, J.; Wang, Z.Y.; Wang, A.; Wang, X.; Wang, J.; Wei, M.; Wides, R.; Xiao, C.; Yan, C.; Yao, A.; Ye, J.; Zhan, M.; Zhang, W.; Zhang, H.; Zhao, Q.; Zheng, L.; Zhong, F.; Zhong, W.; Zhu, S.C.; Zhao, S.; Gilbert, D.; Baumhueter, S.; Spier, G.; Carter, C.; Cravchik, A.; Woodage, T.; Ali, F.; An, H.; Awe, A.; Baldwin, D.; Baden, H.; Barnstead, M.; Barrow, I.; Beeson, K.; Busam, D.; Carver, A.; Center, A.; Cheng, M.L.; Curry, L.; Danaher, S.; Davenport, L.; Desilets, R.; Dietz, S.; Dodson, K.; Doup, L.; Ferriera, S.; Garg, N.; Gluecksmann, A.; Hart, B.; Haynes, J.; Haynes, C.; Heiner, C.; Hladun, S.; Hostin, D.; Houck, J.; Howland, T.; Ibegwam, C.; Johnson, J.; Kalush, F.; Kline, L.; Koduru, S.; Love, A.; Mann, F.; May, D.; McCawley, S.; McIntosh, T.; McMullen, I.; Moy, M.; Moy, L.; Murphy, B.; Nelson, K.; Pfannkoch, C.; Pratts, E.; Puri, V.; Qureshi, H.; Reardon, M.; Rodriguez, R.; Rogers, Y.; Romblad, D.; Ruhfel, B.; Scott, R.; Sitter, C.; Smallwood, M.; Stewart, E.; Strong, R.; Suh, E.; Thomas, R.; Tint, N.N.; Tse, S.; Vech, C.; Wang, G.; Wetter, J.; Williams, S.; Williams, M.; Windsor, S.; Winn-Deen, E.; Wolfe, K.; Zaveri, J.; Zaveri, K.; Abril, J.F.; Guigó, R.; Campbell, M.J.; Sjolander, K.V.; Karlak, B.; Kejariwal, A.; Mi, H.; Lazareva, B.; Hatton, T.; Narechania, A.; Diemer, K.; Muruganujan, A.; Guo, N.; Sato, S.; Bafna, V.; Istrail, S.; Lippert, R.; Schwartz, R.; Walenz, B.; Yooseph, S.; Allen, D.; Basu, A.; Baxendale, J.; Blick, L.; Caminha, M.; Carnes-Stine, J.; Caulk, P.; Chiang, Y.; Coyne, M.; Dahlke, C.; Mays, A.D.; Dombroski, M.; Donnelly, M.; Ely, D.; Esparham, S.; Fosler, C.; Gire, H.; Glanowski, S.; Glasser, K.; Glodek, A.; Gorokhov, M.; Graham, K.; Gropman, B.; Harris, M.; Heil, J.; Henderson, S.; Hoover, J.; Jennings, D.; Jordan, C.; Jordan, J.; Kasha, J.; Kagan, L.; Kraft, C.; Levitsky, A.; Lewis, M.; Liu, X.; Lopez, J.; Ma, D.; Majoros, W.; McDaniel, J.; Murphy, S.; Newman, M.; Nguyen, T.; Nguyen, N.; Nodell, M.; Pan, S.; Peck, J.; Peterson, M.; Rowe, W.; Sanders, R.; Scott, J.; Simpson, M.; Smith, T.; Sprague, A.; Stockwell, T.; Turner, R.; Venter, E.; Wang, M.; Wen, M.; Wu, D.; Wu, M.; Xia, A.; Zandieh, A.; Zhu, X. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar]
- Ressier, K.J.; Sullivan, S.L.; Buck, L.B. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 1993, 73, 597–609. [Google Scholar]
- Vassar, R.; Ngai, J.; Axel, R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 1993, 74, 309–318. [Google Scholar]
- Iwema, C.L.; Fang, H.; Kurtz, D.B.; Youngentob, S.L.; Schwob, J.E. Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J. Neurosci. 2004, 24, 356–369. [Google Scholar]
- Miyamichi, K.; Serizawa, S.; Kimura, H.M.; Sakano, H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J. Neurosci. 2005, 25, 3586–3592. [Google Scholar]
- Norlin, E.M.; Alenius, M.; Gussing, F.; Hagglund, M.; Vedin, V.; Bohm, S. Evidence for gradients of gene expression correlating with zonal topography of the olfactory sensory map. Mol. Cell Neurosci. 2001, 18, 283–295. [Google Scholar]
- Koshimoto, H.; Katoh, K.; Yoshihara, Y.; Nemoto, Y.; Mori, K. Immunohistochemical demonstration of embryonic expression of an odor receptor protein and its zonal distribution in the rat olfactory epithelium. Neurosci. Lett. 1994, 169, 73–76. [Google Scholar]
- Menco, B.P.; Cunningham, A.M.; Qasba, P.; Levy, N.; Reed, R.R. Putative odour receptors localize in cilia of olfactory receptor cells in rat and mouse: a freeze-substitution ultrastructural study. J. Neurocytol 1997, 26, 691–706. [Google Scholar]
- Menco, B.P.; Jackson, J.E. Neuron-like cells on the apical surface of the developing rat olfactory epithelium. Neurosci. Lett. 1997, 239, 117–120. [Google Scholar]
- Menco, B.P.; Jackson, J.E. Cells resembling hair cells in developing rat olfactory and nasal respiratory epithelia. Tissue Cell 1997, 29, 707–713. [Google Scholar]
- Senhupta, P.; Chou, J.H.; Bargmann, C.I. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 1996, 84, 899–909. [Google Scholar]
- Krautwurst, D.; Yau, K.W.; Reed, R.R. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 1998, 95, 917–926. [Google Scholar]
- Zhao, H.; Ivic, L.; Otaki, J.M.; Hashimoto, M.; Mikoshiba, K.; Firestein, S. Functinal expression of a mammalian odorant receptor. Science 1998, 279, 237–241. [Google Scholar]
- Leinders-Zufall, T.; Lane, A.P.; Puche, A.C.; Ma, W.; Novotny, M.V.; Shipley, M.T.; Zufall, F. Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature 2000, 405, 792–796. [Google Scholar]
- Dulac, C.; Axel, R. A novel family of genes encoding putative pheromone receptors in mammals. Cell 1995, 83, 195–206. [Google Scholar]
- Herrada, G.; Dulac, C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell 1997, 90, 763–773. [Google Scholar]
- Matsunami, H.; Buck, L.B. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell 1997, 90, 775–784. [Google Scholar]
- Ryba, N.J.; Tirindelli, R. A new multigene family of putative pheromone receptors. Neuron 1997, 19, 371–379. [Google Scholar]
- Juilfs, D.M.; Fulle, H.J.; Zhao, A.Z.; Houslay, M.D.; Garbers, D.L.; Beavo, J.A. A subset of olfactory neurons that selectively express cGMP-stimulated phosphodiesterase (PDE2) and guanylyl cyclase-D define a unique olfactory signal transduction pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 3388–3395. [Google Scholar]
- Meyer, M.R.; Angele, A.; Kremmer, E.; Kaupp, U.B.; Muller, F. A cGMP-signaling pathway in a subset of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 2000, 97, 10595–10600. [Google Scholar]
- Fulle, H.-J.; Vassar, R.; Foster, D.C.; Yang, R.-B.; Axel, R.; Garbers, D.L. A receptor guanylyl cyclase expressed specifically in olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 1995, 92, 3571–3575. [Google Scholar]
- Palczewski, K.; Subbaraya, I.; Gorczyca, W.A.; Helekar, B.S.; Ruiz, C.C.; Ohguro, H.; Huang, J.; Zhao, X.; Crabb, J.W.; Johnson, R.S.; Walsh, K.A.; Gray-Keller, M.P.; Detwiler, P.B.; Wolfgang Baehr, W. Molecular Cloning and Characterization of Retinal Photoreceptor Guanylyl Cyclase-Activating Protein. Neuron 1994, 13, 395–404. [Google Scholar]
- Moon, C.; Jaberi, P.; Otto-Bruc, A.; Baehr, W.; Palczewski, K.; Ronnett, G.V. Calcium-sensitive particulate guanylyl cyclase as a modulator of cAMP in olfactory receptor neurons. J. Neurosci. 1998, 18, 3195–3205. [Google Scholar]
- Godfrey, P.A.; Malnic, B.; Buck, L.B. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2156–2161. [Google Scholar]
- Mombaerts, P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004, 5, 263–278. [Google Scholar]
- Ressler, K.J.; Sullivan, S.L.; Buck, L.B. Information Coding in the Olfactory System: Evidence for a Stereotyped and Highly Organized Epitope Map in the Olfactory Bulb. Cell 1994, 79, 1245–1255. [Google Scholar]
- Mombaerts, P.; Wang, F.; Dulac, C.; Chao, S.K.; Nemes, A.; Mendelsohn, M.; Edmondson, J.; Axel, R. Visualizing an olfactory sensory map. Cell 1996, 87, 675–686. [Google Scholar]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar]
- Gao, Q.; Chess, A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 1999, 60, 31–39. [Google Scholar]
- Vosshall, L.B.; Amrein, H.; Morozov, P.S.; Rzhetsky, A.; Axel, R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 1999, 96, 725–736. [Google Scholar]
- Shanbhag, S.R.; Muller, B.; Steinbrecht, R.A. Atlas of olfactory organs of Drosophila melanogaster 2. Internal organization and cellular architecture of olfactory sensilla. Arthropod. Struct. Dev. 2000, 29, 211–229. [Google Scholar]
- Goldman, A.L.; Van der Goes van Naters, W.; Lessing, D.; Warr, C.G.; Carlson, J.R. Coexpression of two functional odor receptors in one neuron. Neuron 2005, 45, 661–666. [Google Scholar]
- de Bruyne, M.; Foster, K.; Carlson, J.R. Odor coding in the Drosophila antenna. Neuron 2001, 30, 537–552. [Google Scholar]
- Hallem, E.A.; Ho, M.G.; Carlson, J.R. The molecular basis of odor coding in the Drosophila antenna. Cell 2004, 117, 965–979. [Google Scholar]
- van der Goes van Naters, W.; Carlson, J.R. Receptors and neurons for fly odors in Drosophila. Curr. Biol. 2007, 17, 606–612. [Google Scholar]
- Yao, C.A.; Ignell, R.; Carlson, J.R. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J. Neurosci. 2005, 25, 8359–8367. [Google Scholar]
- Gao, Q.; Yuan, B.; Chess, A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat. Neurosci. 2000, 3, 780–785. [Google Scholar]
- Vosshall, L.B.; Wong, A.M.; Axel, R. An olfactory sensory map in the fly brain. Cell 2000, 102, 147–159. [Google Scholar]
- Aceves-Pina, E.O.; Quinn, W.G. Learning in Normal and Mutant Drosophila Larvae. Science 1979, 206, 93–96. [Google Scholar]
- Monte, P.; Woodard, C.; Ayer, R.; Lilly, M.; Sun, H.; Carlson, J. Characterization of the larval olfactory response in Drosophila and its genetic basis. Behav. Genet. 1989, 19, 267–283. [Google Scholar]
- Python, F.; Stocker, R.F. Adult-like complexity of the larval antennal lobe of D. melanogaster despite markedly low numbers of odorant receptor neurons. J. Comp. Neurol. 2002, 445, 374–387. [Google Scholar]
- Kreher, S.A.; Kwon, J.Y.; Carlson, J.R. The molecular basis of odor coding in the Drosophila larva. Neuron 2005, 46, 445–456. [Google Scholar]
- Troemel, E.R.; Chou, J.H.; Dwyer, N.D.; Colbert, H.A.; Bargmann, C.I. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 1995, 83, 207–218. [Google Scholar]
- Sato, K.; Pellegrino, M.; Nakagawa, T.; Nakagawa, T.; Vosshall, L.B.; Touhara, K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 2008, 452, 1002–1006. [Google Scholar]
- Wicher, D.; Schafer, R.; Bauernfeind, R.; Stensmyr, M.C.; Heller, R.; Heinemann, S.H.; Hansson, B.S. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 2008, 452, 1007–1011. [Google Scholar]
- Couto, A.; Alenius, M.; Dickson, B.J. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 2005, 15, 1535–1547. [Google Scholar]
- Larsson, H.P.; Kleene, S.J.; Lecar, H. Noise analysis of ion channels in non-space-clamped cables: estimates of channel parameters in olfactory cilia. Biophys. J. 1997, 72, 1193–1203. [Google Scholar]
- Serizawa, S.; Miyamichi, K.; Nakatani, H.; Suzuki, M.; Saito, M.; Yoshihara, Y.; Sakano, H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 2003, 302, 2088–2094. [Google Scholar]
- Benton, R.; Sachse, S.; Michnick, S.W.; Vosshall, L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006, 4, e20. [Google Scholar]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100 Suppl 2, 14537–14542. [Google Scholar]
- Dobritsa, A.A.; van der Goes van Naters, W.; Warr, C.G.; Steinbrecht, R.A.; Carlson, J.R. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 2003, 37, 827–841. [Google Scholar]
- Fishilevich, E.; Domingos, A.I.; Asahina, K.; Naef, F.; Vosshall, L.B.; Louis, M. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 2005, 15, 2086–2096. [Google Scholar]
- Jones, W.D.; Nguyen, T.A.; Kloss, B.; Lee, K.J.; Vosshall, L.B. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 2005, 15, R119–121. [Google Scholar]
- Neuhaus, E.M.; Gisselmann, G.; Zhang, W.; Dooley, R.; Stortkuhl, K.; Hatt, H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 2005, 8, 15–17. [Google Scholar]
- Hallem, E.A.; Carlson, J.R. Coding of odors by a receptor repertoire. Cell 2006, 125, 143–160. [Google Scholar]
- Hallem, E.A.; Carlson, J.R. The odor coding system of Drosophila. Trends. Genet. 2004, 20, 453–459. [Google Scholar]
- Hallem, E.A.; Carlson, J.R. The spatial code for odors is changed by conditioning. Neuron 2004, 42, 359–361. [Google Scholar]
- Ha, T.S.; Smith, D.P. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 2006, 26, 8727–8733. [Google Scholar]
- Kurtovic, A.; Widmer, A.; Dickson, B.J. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 2007, 446, 542–546. [Google Scholar]
- Zawistowski, C.A.; DeVita, M.A. Non-heartbeating organ donation: a review. J. Intensive Care Med. 2003, 18, 189–197. [Google Scholar]
- Laughlin, J.D.; Ha, T.S.; Jones, D.N.; Smith, D.P. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 2008, 133, 1255–1265. [Google Scholar]
- Zwiebel, L.J.; Takken, W. Olfactory regulation of mosquito-host interactions. Insect Biochem. Mol. Biol. 2004, 34, 645–652. [Google Scholar]
- Suh, G.S.; Wong, A.M.; Hergarden, A.C.; Wang, J.W.; Simon, A.F.; Benzer, S.; Axel, R.; Anderson, D.J. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature 2004, 431, 854–859. [Google Scholar]
- Clyne, P.J.; Warr, C.G.; Carlson, J.R. Candidate taste receptors in Drosophila. Science 2000, 287, 1830–1834. [Google Scholar]
- Jones, W.D.; Cayirlioglu, P.; Kadow, I.G.; Vosshall, L.B. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 2007, 445, 86–90. [Google Scholar]
- Kwon, J.Y.; Dahanukar, A.; Weiss, L.A.; Carlson, J.R. The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. USA 2007, 104, 3574–3578. [Google Scholar]
- Hill, C.A.; Fox, A.N.; Pitts, R.J.; Kent, L.B.; Tan, P.L.; Chrystal, M.A.; Cravchik, A.; Collins, F.H.; Robertson, H.M.; Zwiebel, L.J. G protein-coupled receptors in Anopheles gambiae. Science 2002, 298, 176–178. [Google Scholar]
- Lu, T.; Qiu, Y.T.; Wang, G.; Kwon, J.Y.; Rutzler, M.; Kwon, H.W.; Pitts, R.J.; van Loon, J.J.; Takken, W.; Carlson, J.R.; Zwiebel, L.J. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 2007, 17, 1533–1544. [Google Scholar]







© 2008 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Song, H.-G.; Young Kwon, J.; Soo Han, H.; Bae, Y.-C.; Moon, C. First Contact to Odors: Our Current Knowledge about Odorant Receptor. Sensors 2008, 8, 6303-6320. https://doi.org/10.3390/s8106303
Song H-G, Young Kwon J, Soo Han H, Bae Y-C, Moon C. First Contact to Odors: Our Current Knowledge about Odorant Receptor. Sensors. 2008; 8(10):6303-6320. https://doi.org/10.3390/s8106303
Chicago/Turabian StyleSong, Hyoung-Gon, Jae Young Kwon, Hyung Soo Han, Yong-Chul Bae, and Cheil Moon. 2008. "First Contact to Odors: Our Current Knowledge about Odorant Receptor" Sensors 8, no. 10: 6303-6320. https://doi.org/10.3390/s8106303
APA StyleSong, H. -G., Young Kwon, J., Soo Han, H., Bae, Y. -C., & Moon, C. (2008). First Contact to Odors: Our Current Knowledge about Odorant Receptor. Sensors, 8(10), 6303-6320. https://doi.org/10.3390/s8106303