Progress Report: Antimicrobial Drug Discovery in the Resistance Era
Abstract
:1. Introduction
2. Selection of Literature
3. Antimicrobial Drug Discovery: Timeline
4. Historical Aspects for the Development of Antimicrobial Resistance
5. Antimicrobial Peptides as New Molecules against Drug Resistance
6. New Antimicrobial Drug Targets
6.1. NagZ Protein
6.2. AmpG Protein
6.3. Polyphosphate Kinase
6.4. Cytochrome bc1 Complex
7. Recent Developments in Drug Discovery against Drug Resistance
7.1. Drug Screening and Non-Conventional Growth Media
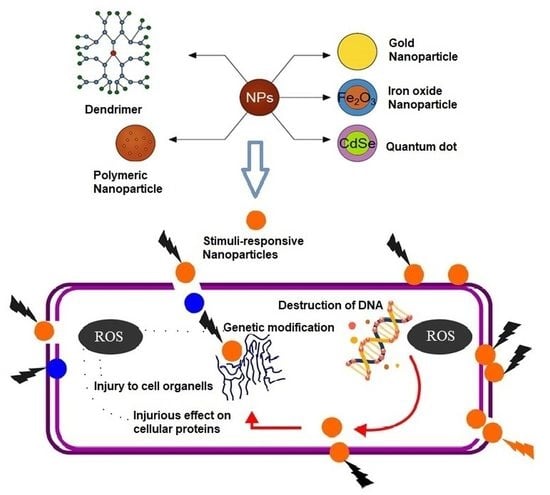
7.2. Nanomaterials for the Design and Delivery of Antimicrobial Agents
8. Antibiotic Adjuvants and Combination Therapies
9. Challenges and Future Outlook
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMP | Antimicrobial peptide |
| AMR | Antimicrobial resistance |
| anhMurNAc | 1,6-anhydroumuropeptides |
| ARGs | Antibiotic resistance genes |
| ARP | Antibiotic resistance platform |
| ATP | Adenosine triphosphate |
| BrCND | Brominated carbon nanodot |
| CDC | Center for disease control and prevention |
| CRE | Carbapenem-resistant Enterobacterales |
| ESKAPE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species |
| FA | Fatty acid |
| FDA | Food and drug administration |
| GLASS | Global antimicrobial resistance and surveillance |
| GlcNAc | N-acetylglucosaminyl |
| GSK | GalaxoSmithKline |
| MD | Molecular dynamic |
| MDR | Multi-drug resistance |
| MIP | Molecularly imprinted polymer |
| MM-GBSA | Molecular-generalized born surface area |
| MMV | Medicine for malaria venture |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MTB | Multi-drug resistant tuberculosis |
| NIPs | Non-molecularly imprinted polymers |
| O2M | Overlap2 method |
| PG | Peptidoglycans |
| PolyP | Polyphosphates |
| PPK | Polyphosphate kinase |
| QSAR | Quantitative structure-activity relationship |
| SAR | Structure-activity relationship |
| SDGs | Sustainable development goals |
| UV | Ultraviolet |
| VRSA | Vancomycin-resistant staphylococcus aureus |
| WHO | World health organization |
| XDR | Extensively drug-resistant |
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 18 March 2022).
- Liu, Y.; Tong, Z.; Shi, J.; Li, R.; Upton, M.; Wang, Z. Drug repurposing for next-generation combination therapies against multidrug-resistant bacteria. Theranostics 2021, 11, 4910–4928. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 26 January 2022).
- Cardoso, P.; Glossop, H.; Meikle, T.G.; Aburto-Medina, A.; Conn, C.E.; Sarojini, V.; Valery, C. Molecular engineering of antimicrobial peptides: Microbial targets, peptide motifs and translation opportunities. Biophys. Rev. 2021, 13, 35–69. [Google Scholar] [CrossRef] [PubMed]
- Coates, M.M.; Kintu, A.; Gupta, N.; Wroe, E.B.; Adler, A.J.; Kwan, G.F.; Park, P.H.; Rajbhandari, R.; Byrne, A.L.; Casey, D.C.; et al. Burden of non-communicable diseases from infectious causes in 2017: A modelling study. Lancet. Glob. Health 2020, 8, e1489–e1498. [Google Scholar] [CrossRef]
- Buckley, B.S.; Henschke, N.; Bergman, H.; Skidmore, B.; Klemm, E.J.; Villanueva, G.; Garritty, C.; Paul, M. Impact of vaccination on antibiotic usage: A systematic review and meta-analysis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2019, 25, 1213–1225. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Urbán, E.; Stájer, A.; Baráth, Z. Antimicrobial Resistance in the Context of the Sustainable Development Goals: A Brief Review. Eur. J. Investig. Health Psychol. Educ. 2021, 11, 71–82. [Google Scholar] [CrossRef]
- Marquette, A.; Bechinger, B. Biophysical Investigations Elucidating the Mechanisms of Action of Antimicrobial Peptides and Their Synergism. Biomolecules 2018, 8, 18. [Google Scholar] [CrossRef] [Green Version]
- Hobson, C.; Chan, A.N.; Wright, G.D. The Antibiotic Resistome: A Guide for the Discovery of Natural Products as Antimicrobial Agents. Chem. Rev. 2021, 121, 3464–3494. [Google Scholar] [CrossRef]
- Anup, N.; Chavan, T.; Chavan, S.; Polaka, S.; Kalyane, D.; Abed, S.N.; Venugopala, K.N.; Kalia, K.; Tekade, R.K. Reinforced electrospun nanofiber composites for drug delivery applications. J. Biomed. Mater. Res. Part A 2021, 109, 2036–2064. [Google Scholar] [CrossRef]
- Parish, T. Steps to address anti-microbial drug resistance in today’s drug discovery. Expert Opin. Drug Discov. 2019, 14, 91–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munir, M.U.; Ahmad, M.M. Nanomaterials Aiming to Tackle Antibiotic-Resistant Bacteria. Pharmaceutics 2022, 14, 582. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.Y.; Ko, W.C.; Hsueh, P.R. Nanoparticles in the Treatment of Infections Caused by Multidrug-Resistant Organisms. Front. Pharmacol. 2019, 10, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.; Vishwakarma, K.; Ramawat, N.; Rai, P.; Singh, V.K.; Mishra, R.K.; Kumar, V.; Tripathi, D.K.; Sharma, S. Nanomaterials and microbes' interactions: A contemporary overview. 3 Biotech 2019, 9, 68. [Google Scholar] [CrossRef]
- Blaskovich, M.A.; Butler, M.S.; Cooper, M.A. Polishing the tarnished silver bullet: The quest for new antibiotics. Essays Biochem. 2017, 61, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Haroun, M.; Tratrat, C.; Petrou, A.; Geronikaki, A.; Ivanov, M.; Ćirić, A.; Soković, M.; Nagaraja, S.; Venugopala, K.N.; Balachandran Nair, A.; et al. Exploration of the antimicrobial effects of benzothiazolylthiazolidin-4-one and in silico mechanistic investigation. Molecules 2021, 26, 4061. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Khulan, A.; Kim, J. Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 2019, 9, 6666. [Google Scholar] [CrossRef]
- Singh, V.; Haque, S.; Singh, H.; Verma, J.; Vibha, K.; Singh, R.; Jawed, A.; Tripathi, C.K. Isolation, Screening, and Identification of Novel Isolates of Actinomycetes from India for Antimicrobial Applications. Front. Microbiol. 2016, 7, 1921. [Google Scholar] [CrossRef] [Green Version]
- Van Goethem, M.W.; Makhalanyane, T.P.; Cowan, D.A.; Valverde, A. Cyanobacteria and Alphaproteobacteria May Facilitate Cooperative Interactions in Niche Communities. Front. Microbiol. 2017, 8, 2099. [Google Scholar] [CrossRef]
- Behie, S.W.; Bonet, B.; Zacharia, V.M.; McClung, D.J.; Traxler, M.F. Molecules to Ecosystems: Actinomycete Natural Products In situ. Front. Microbiol. 2016, 7, 2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef] [PubMed]
- Pankhurst, C.E. Symbiotic effectiveness of antibiotic-resistant mutants of fast- and slow-growing strains of Rhizobium nodulating Lotus species. Can. J. Microbiol. 1977, 23, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Fiedler, H.P. A guide to successful bioprospecting: Informed by actinobacterial systematics. Antonie Van Leeuwenhoek 2010, 98, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [Green Version]
- Hosaka, T.; Ohnishi-Kameyama, M.; Muramatsu, H.; Murakami, K.; Tsurumi, Y.; Kodani, S.; Yoshida, M.; Fujie, A.; Ochi, K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009, 27, 462–464. [Google Scholar] [CrossRef]
- Wright, G.D.; Sutherland, A.D. New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 2007, 13, 260–267. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, S.; Shen, J.; Zhu, K. Nonribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat. Prod. Rep. 2019, 36, 573–592. [Google Scholar] [CrossRef] [Green Version]
- Antibiotics Special Issue: Challenges and Opportunities in Antibiotic Discovery and Development. ACS Infect. Dis. 2020, 6, 1286–1288. [CrossRef]
- Morsy, M.A.; Ali, E.M.; Kandeel, M.; Venugopala, K.N.; Nair, A.B.; Greish, K.; El-Daly, M. Screening and molecular docking of novel benzothiazole derivatives as potential antimicrobial agents. Antibiotics 2020, 9, 221. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Uppar, V.; Chandrashekharappa, S.; Abdallah, H.H.; Pillay, M.; Deb, P.K.; Morsy, M.A.; Aldhubiab, B.E.; Attimarad, M.; Nair, A.B.; et al. Cytotoxicity and antimycobacterial properties of pyrrolo[1,2-a]quinoline derivatives: Molecular target identification and molecular docking studies. Antibiotics 2020, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet. Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- Steenbergen, J.N.; Alder, J.; Thorne, G.M.; Tally, F.P. Daptomycin: A lipopeptide antibiotic for the treatment of serious Gram-positive infections. J. Antimicrob. Chemother. 2005, 55, 283–288. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.K.; Abramovitch, R.B. Small Molecules That Sabotage Bacterial Virulence. Trends Pharmacol. Sci. 2017, 38, 339–362. [Google Scholar] [CrossRef] [Green Version]
- Dickey, S.W.; Cheung, G.Y.C.; Otto, M. Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017, 16, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Worthington, R.J.; Melander, C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013, 31, 177–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, R.; Xiao, X.; Wang, Z. Antibiotic adjuvants: An alternative approach to overcome multi-drug resistant Gram-negative bacteria. Crit. Rev. Microbiol. 2019, 45, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Peyclit, L.; Baron, S.A.; Rolain, J.M. Drug Repurposing to Fight Colistin and Carbapenem-Resistant Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 193. [Google Scholar] [CrossRef]
- Peraman, R.; Sure, S.K.; Dusthackeer, V.N.A.; Chilamakuru, N.B.; Yiragamreddy, P.R.; Pokuri, C.; Kutagulla, V.K.; Chinni, S. Insights on recent approaches in drug discovery strategies and untapped drug targets against drug resistance. Future J. Pharm. Sci. 2021, 7, 56. [Google Scholar] [CrossRef]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar] [CrossRef] [Green Version]
- Ganz, T.; Lehrer, R.I. Defensins. Curr. Opin. Immunol. 1994, 6, 584–589. [Google Scholar] [CrossRef]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The interaction of antimicrobial peptides with membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016, 24, 862–871. [Google Scholar] [CrossRef]
- Brown, D. Antibiotic resistance breakers: Can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 2015, 14, 821–832. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Chandrashekharappa, S.; Pillay, M.; Abdallah, H.H.; Mahomoodally, F.M.; Bhandary, S.; Chopra, D.; Attimarad, M.; Aldhubiab, B.E.; Nair, A.B.; et al. Computational, crystallographic studies, cytotoxicity and anti-tubercular activity of substituted 7-methoxy-indolizine analogues. PLoS ONE 2019, 14, e0217270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Naghavi, M. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Crofts, T.S.; Gasparrini, A.J.; Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 2017, 15, 422–434. [Google Scholar] [CrossRef] [Green Version]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Singh, V.A.; Pottathil, S. Metallo-β-lactamase- and serine carbapenemase-producing Klebsiella spp.: A global challenge. J. Glob. Antimicrob. Resist. 2018, 12, 185–186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Singh, V.A.; Beniwal, V.; Pottathil, S. Modified Carba NP Test: Simple and rapid method to differentiate KPC- and MBL-producing Klebsiella species. J. Clin. Lab. Anal. 2018, 32, e22448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinu, P.; Bareja, R.; Nair, A.B.; Mishra, V.; Hussain, S.; Venugopala, K.N.; Sreeharsha, N.; Attimarad, M.; Rasool, S.T. Monitoring of Non-β-Lactam Antibiotic Resistance-Associated Genes in ESBL Producing Enterobacterales Isolates. Antibiotics 2020, 9, 884. [Google Scholar] [CrossRef] [PubMed]
- Hammadi, A.; Aga, Q.; Nimer, N.; Shinu, P.; Nair, A. Antimicrobial resistance and presence of Class 1 integrons in Pseudomonas aeruginosa isolates from burn and wound infections. J. Pharm. Negat. Result 2020, 11, 19–22. [Google Scholar]
- Stephenson, B.; Lanzas, C.; Lenhart, S.; Ponce, E.; Bintz, J.; Dubberke, E.R.; Day, J. Comparing intervention strategies for reducing Clostridioides difficile transmission in acute healthcare settings: An agent-based modeling study. BMC Infect. Dis. 2020, 20, 799. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Shinu, P.; Singh, V.; Nair, A. Isoniazid and rifampin drug susceptibility testing: Application of 2,3,5-triphenyl tetrazolium chloride assay and microscopic-observation drug-susceptibility assay directly on Ziehl-Neelsen smear positive sputum specimens. Braz. J. Infect. Dis. 2016, 20, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Shinu, P.; Nair, A.B.; Hussain, S.; Morsy, M.A.; Soliman, W.E. Pancreatin-cetyl pyridinium chloride digestion and decontamination method; a novel, sensitive, cost-effective method for culturing mycobacterium tuberculosis. Microorganisms 2021, 9, 2025. [Google Scholar] [CrossRef]
- Shinu, P.; Singh, V.A.; Nair, A.; Venugopala, K.N.; Akrawi, S.H. Papain-cetylpyridinium chloride and pepsin-cetylpyridinium chloride; two novel, highly sensitive, concentration, digestion and decontamination techniques for culturing mycobacteria from clinically suspected pulmonary tuberculosis cases. PLoS ONE 2020, 15, e0236700. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2021. Available online: https://www.who.int/publications/i/item/9789240027336 (accessed on 26 January 2022).
- Shinu, P.; Morsy, M.A.; Deb, P.K.; Nair, A.B.; Goyal, M.; Shah, J.; Kotta, S. SARS CoV-2 Organotropism Associated Pathogenic Relationship of Gut-Brain Axis and Illness. Front. Mol. Biosci. 2020, 7, 439. [Google Scholar] [CrossRef]
- Hussain, S.; Rasool, S.T.; Pottathil, S. The Evolution of Severe Acute Respiratory Syndrome Coronavirus-2 during Pandemic and Adaptation to the Host. J. Mol. Evol. 2021, 89, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, Y.; Song, Z.; Tan, Z.; Cheng, J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv. Drug Deliv. Rev. 2021, 170, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef] [Green Version]
- Mangoni, M.L.; Casciaro, B. Development of Antimicrobial Peptides from Amphibians. Antibiotics 2020, 9, 772. [Google Scholar] [CrossRef]
- Selsted, M.E.; Novotny, M.J.; Morris, W.L.; Tang, Y.Q.; Smith, W.; Cullor, J.S. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 1992, 267, 4292–4295. [Google Scholar] [CrossRef]
- Mojsoska, B.; Jenssen, H. Peptides and Peptidomimetics for Antimicrobial Drug Design. Pharmaceuticals 2015, 8, 366–415. [Google Scholar] [CrossRef]
- Kiho, T.; Yoshida, I.; Katsuragawa, M.; Sakushima, M.; Usui, S.; Ukai, S. Polysaccharides in Fungi. XXXIV. A polysaccharide from the fruiting bodies of Amanita muscaria and the antitumor activity of its carboxymethylated product. Biol. Pharm. Bull. 1994, 17, 1460–1462. [Google Scholar] [CrossRef] [Green Version]
- Wiater, A.; Paduch, R.; Pleszczyńska, M.; Próchniak, K.; Choma, A.; Kandefer-Szerszeń, M.; Szczodrak, J. α-(1 → 3)-D-glucans from fruiting bodies of selected macromycetes fungi and the biological activity of their carboxymethylated products. Biotechnol. Lett. 2011, 33, 787–795. [Google Scholar] [CrossRef] [Green Version]
- Harder, J.; Bartels, J.; Christophers, E.; Schroder, J.M. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 2001, 276, 5707–5713. [Google Scholar] [CrossRef] [Green Version]
- Savoia, D.; Guerrini, R.; Marzola, E.; Salvadori, S. Synthesis and antimicrobial activity of dermaseptin S1 analogues. Bioorganic Med. Chem. 2008, 16, 8205–8209. [Google Scholar] [CrossRef] [Green Version]
- Lai, R.; Zheng, Y.T.; Shen, J.H.; Liu, G.J.; Liu, H.; Lee, W.H.; Tang, S.Z.; Zhang, Y. Antimicrobial peptides from skin secretions of Chinese red belly toad Bombina maxima. Peptides 2002, 23, 427–435. [Google Scholar] [CrossRef]
- Meng, H.; Kumar, K. Antimicrobial activity and protease stability of peptides containing fluorinated amino acids. J. Am. Chem. Soc. 2007, 129, 15615–15622. [Google Scholar] [CrossRef] [PubMed]
- Kapil, S.; Sharma, V. d-Amino acids in antimicrobial peptides: A potential approach to treat and combat antimicrobial resistance. Can. J. Microbiol. 2021, 67, 119–137. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, Z.; Cao, S.; Wang, H.; Jiang, C.; Hussain, M.A.; Pang, S. Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine α(S1)-Casein. Molecules 2018, 23, 1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wade, D.; Boman, A.; Wåhlin, B.; Drain, C.M.; Andreu, D.; Boman, H.G.; Merrifield, R.B. All-D amino acid-containing channel-forming antibiotic peptides. Proc. Natl. Acad. Sci. USA 1990, 87, 4761–4765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oren, Z.; Shai, Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: Structure-function study. Biochemistry 1997, 36, 1826–1835. [Google Scholar] [CrossRef] [PubMed]
- Narayanaswamy, V.K.; Albericio, F.; Coovadia, Y.M.; Kruger, H.G.; Maguire, G.E.; Pillay, M.; Govender, T. Total synthesis of a depsidomycin analogue by convergent solid-phase peptide synthesis and macrolactonization strategy for antitubercular activity. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2011, 17, 683–689. [Google Scholar] [CrossRef]
- Teixeira, M.C.; Carbone, C.; Sousa, M.C.; Espina, M.; Garcia, M.L.; Sanchez-Lopez, E.; Souto, E.B. Nanomedicines for the Delivery of Antimicrobial Peptides (AMPs). Nanomaterials 2020, 10, 560. [Google Scholar] [CrossRef] [Green Version]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, e24122. [Google Scholar] [CrossRef]
- Huang, Y.W.; Wu, C.J.; Hu, R.M.; Lin, Y.T.; Yang, T.C. Interplay among membrane-bound lytic transglycosylase D1, the CreBC two-component regulatory system, the AmpNG-AmpDI-NagZ-AmpR regulatory circuit, and L1/L2 β-lactamase expression in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2015, 59, 6866–6872. [Google Scholar] [CrossRef] [Green Version]
- Acebrón, I.; Mahasenan, K.V.; De Benedetti, S.; Lee, M.; Artola-Recolons, C.; Hesek, D.; Wang, H.; Hermoso, J.A.; Mobashery, S. Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa. J. Am. Chem. Soc. 2017, 139, 6795–6798. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, C.; Chen, Y.; Hao, H.; Liang, J.; Duan, R.; Guo, Z.; Zhang, J.; Zhao, Z.; Jing, H.; et al. Role of Low-Molecular-Mass Penicillin-Binding Proteins, NagZ and AmpR in AmpC β-lactamase Regulation of Yersinia enterocolitica. Front. Cell. Infect. Microbiol. 2017, 7, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.W.; Wang, Y.; Lin, Y.; Lin, C.; Lin, Y.T.; Hsu, C.C.; Yang, T.C. Impacts of Penicillin Binding Protein 2 Inactivation on β-Lactamase Expression and Muropeptide Profile in Stenotrophomonas maltophilia. mSystems 2017, 2, e00077-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhoopalan, S.V.; Piekarowicz, A.; Lenz, J.D.; Dillard, J.P.; Stein, D.C. nagZ Triggers Gonococcal Biofilm Disassembly. Sci. Rep. 2016, 6, 22372. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zeng, J.; Zhou, Q.; Yu, X.; Zhong, Y.; Wang, F.; Du, H.; Nie, F.; Pang, X.; Wang, D.; et al. Elevating NagZ Improves Resistance to β-Lactam Antibiotics via Promoting AmpC β-Lactamase in Enterobacter cloacae. Front. Microbiol. 2020, 11, 586729. [Google Scholar] [CrossRef]
- Dik, D.A.; Fisher, J.F.; Mobashery, S. Cell-Wall Recycling of the Gram-Negative Bacteria and the Nexus to Antibiotic Resistance. Chem. Rev. 2018, 118, 5952–5984. [Google Scholar] [CrossRef]
- Borisova, M.; Gisin, J.; Mayer, C. The N-Acetylmuramic Acid 6-Phosphate Phosphatase MupP Completes the Pseudomonas Peptidoglycan Recycling Pathway Leading to Intrinsic Fosfomycin Resistance. mBio 2017, 8, e00092-17. [Google Scholar] [CrossRef] [Green Version]
- Torrens, G.; Pérez-Gallego, M.; Moya, B.; Munar-Bestard, M.; Zamorano, L.; Cabot, G.; Blázquez, J.; Ayala, J.A.; Oliver, A.; Juan, C. Targeting the permeability barrier and peptidoglycan recycling pathways to disarm Pseudomonas aeruginosa against the innate immune system. PLoS ONE 2017, 12, e0181932. [Google Scholar] [CrossRef] [Green Version]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 69, 1446–1455. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Ying, J.; Yang, G.; Li, A.; Wang, J.; Lu, J.; Wang, J.; Xu, T.; Yi, H.; Li, K.; et al. Structure-Function Analysis of the Transmembrane Protein AmpG from Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0168060. [Google Scholar] [CrossRef] [Green Version]
- Mallik, D.; Pal, S.; Ghosh, A.S. Involvement of AmpG in mediating a dynamic relationship between serine beta-lactamase induction and biofilm-forming ability of Escherichia coli. FEMS Microbiol. Lett. 2018, 365, fny065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korfmann, G.; Sanders, C.C. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob. Agents Chemother. 1989, 33, 1946–1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mark, B.L.; Vocadlo, D.J.; Oliver, A. Providing β-lactams a helping hand: Targeting the AmpC β-lactamase induction pathway. Future Microbiol. 2011, 6, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Wu, C.; Lin, C.; Li, P.; Zhang, K.; Xu, L.; Liu, Y.; Lu, J.; Cheng, C.; Bao, Q.; et al. The Structure of ampG Gene in Pseudomonas aeruginosa and Its Effect on Drug Resistance. Can. J. Infect. Dis. Med. Microbiol. J. Can. Des Mal. Infect. Et De La Microbiol. Med. 2018, 2018, 7170416. [Google Scholar] [CrossRef] [Green Version]
- Candon, H.L.; Allan, B.J.; Fraley, C.D.; Gaynor, E.C. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 2007, 189, 8099–8108. [Google Scholar] [CrossRef] [Green Version]
- Nocek, B.; Kochinyan, S.; Proudfoot, M.; Brown, G.; Evdokimova, E.; Osipiuk, J.; Edwards, A.M.; Savchenko, A.; Joachimiak, A.; Yakunin, A.F. Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17730–17735. [Google Scholar] [CrossRef] [Green Version]
- Bashatwah, R.M.; Khanfar, M.A.; Bardaweel, S.K. Discovery of potent polyphosphate kinase 1 (PPK1) inhibitors using structure-based exploration of PPK1Pharmacophoric space coupled with docking analyses. J. Mol. Recognit. JMR 2018, 31, e2726. [Google Scholar] [CrossRef]
- Alhaji Isa, M.; Singh Majumdar, R. Computer-aided drug design based on comparative modeling, molecular docking and molecular dynamic simulation of Polyphosphate kinase (PPK) from Mycobacterium tuberculosis. J. Proteins Proteom. 2019, 10, 55–68. [Google Scholar] [CrossRef]
- Singh, M.; Tiwari, P.; Arora, G.; Agarwal, S.; Kidwai, S.; Singh, R. Establishing Virulence Associated Polyphosphate Kinase 2 as a drug target for Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 26900. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.S.; Teo, A.S.; Wong, S.Y. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2001, 45, 2157–2159. [Google Scholar] [CrossRef] [Green Version]
- Cloete, R.; Oppon, E.; Murungi, E.; Schubert, W.D.; Christoffels, A. Resistance related metabolic pathways for drug target identification in Mycobacterium tuberculosis. BMC Bioinform. 2016, 17, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kessl, J.J.; Meshnick, S.R.; Trumpower, B.L. Modeling the molecular basis of atovaquone resistance in parasites and pathogenic fungi. Trends Parasitol. 2007, 23, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, L.; Hu, J.; Duan, H.; Lin, D.; Liu, P.; Meng, Q.; Li, B.; Si, N.; Liu, C.; et al. Resistance Mechanisms and Molecular Docking Studies of Four Novel QoI Fungicides in Peronophythora litchii. Sci. Rep. 2015, 5, 17466. [Google Scholar] [CrossRef] [PubMed]
- Fisher, N.; Meunier, B.; Biagini, G.A. The cytochrome bc(1) complex as an antipathogenic target. FEBS Lett. 2020, 594, 2935–2952. [Google Scholar] [CrossRef] [PubMed]
- Venugopala, K.N.; Ramachandra, P.; Tratrat, C.; Gleiser, R.M.; Bhandary, S.; Chopra, D.; Morsy, M.A.; Aldhubiab, B.E.; Attimarad, M.; Nair, A.B.; et al. Larvicidal activities of 2-aryl-2,3-dihydroquinazolin -4-ones against malaria vector anopheles arabiensis, in silico ADMET prediction and molecular target investigation. Molecules 2020, 25, 1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, R.W.; Kelly, J.X.; Smilkstein, M.J.; Dodean, R.; Hinrichs, D.; Riscoe, M.K. Antimalarial quinolones: Synthesis, potency, and mechanistic studies. Exp. Parasitol. 2008, 118, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Nilsen, A.; LaCrue, A.N.; White, K.L.; Forquer, I.P.; Cross, R.M.; Marfurt, J.; Mather, M.W.; Delves, M.J.; Shackleford, D.M.; Saenz, F.E.; et al. Quinolone-3-diarylethers: A new class of antimalarial drug. Sci. Transl. Med. 2013, 5, 177ra137. [Google Scholar] [CrossRef] [Green Version]
- Chiu, J.E.; Renard, I.; Pal, A.C.; Singh, P.; Vydyam, P.; Thekkiniath, J.; Kumar, M.; Gihaz, S.; Pou, S.; Winter, R.W.; et al. Effective Therapy Targeting Cytochrome bc(1) Prevents Babesia Erythrocytic Development and Protects from Lethal Infection. Antimicrob. Agents Chemother. 2021, 65, e0066221. [Google Scholar] [CrossRef]
- Iacobino, A.; Fattorini, L.; Giannoni, F. Drug-resistant tuberculosis 2020: Where we stand. Appl. Sci. 2020, 10, 2153. [Google Scholar] [CrossRef] [Green Version]
- Spriggs, A.C.; Dakora, F.D. Assessing the suitability of antibiotic resistance markers and the indirect ELISA technique for studying the competitive ability of selected Cyclopia Vent. rhizobia under glasshouse and field conditions in South Africa. BMC Microbiol. 2009, 9, 142. [Google Scholar] [CrossRef] [Green Version]
- Pandey, B.; Grover, S.; Kaur, J.; Grover, A. Analysis of mutations leading to para-aminosalicylic acid resistance in Mycobacterium tuberculosis. Sci. Rep. 2019, 9, 13617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, N.D.; Carey, A.F.; Yang, J.; Zhao, Y.; Fortune, S.M. Bacterial Genome-Wide Association Identifies Novel Factors That Contribute to Ethionamide and Prothionamide Susceptibility in Mycobacterium tuberculosis. mBio 2019, 10, e00616-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perdigão, J.; Portugal, I. Genetics and roadblocks of drug resistant tuberculosis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 72, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, V.; Singh, P.K.; Prakash, S.; Jain, A. Second Line Injectable Drug Resistance and Associated Genetic Mutations in Newly Diagnosed Cases of Multidrug-Resistant Tuberculosis. Microb. Drug Resist. 2020, 26, 971–975. [Google Scholar] [CrossRef]
- Iannazzo, L.; Soroka, D.; Triboulet, S.; Fonvielle, M.; Compain, F.; Dubée, V.; Mainardi, J.L.; Hugonnet, J.E.; Braud, E.; Arthur, M.; et al. Routes of Synthesis of Carbapenems for Optimizing Both the Inactivation of L,D-Transpeptidase LdtMt1 of Mycobacterium tuberculosis and the Stability toward Hydrolysis by β-Lactamase BlaC. J. Med. Chem. 2016, 59, 3427–3438. [Google Scholar] [CrossRef]
- Nusrath Unissa, A.; Hanna, L.E. Molecular mechanisms of action, resistance, detection to the first-line anti tuberculosis drugs: Rifampicin and pyrazinamide in the post whole genome sequencing era. Tuberculosis 2017, 105, 96–107. [Google Scholar] [CrossRef]
- Fujiwara, M.; Kawasaki, M.; Hariguchi, N.; Liu, Y.; Matsumoto, M. Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis 2018, 108, 186–194. [Google Scholar] [CrossRef]
- Chen, X.; He, G.; Wang, S.; Lin, S.; Chen, J.; Zhang, W. Evaluation of Whole-Genome Sequence Method to Diagnose Resistance of 13 Anti-tuberculosis Drugs and Characterize Resistance Genes in Clinical Multi-Drug Resistance Mycobacterium tuberculosis Isolates From China. Front. Microbiol. 2019, 10, 1741. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Xiao, T.Y.; Liu, H.C.; Zhao, X.Q.; Liu, Z.G.; Li, Y.N.; Zeng, H.; Zhao, L.L.; Wan, K.L. Mutations within embCAB Are Associated with Variable Level of Ethambutol Resistance in Mycobacterium tuberculosis Isolates from China. Antimicrob. Agents Chemother. 2018, 62, e01279-17. [Google Scholar] [CrossRef] [Green Version]
- Evangelopoulos, D.; Prosser, G.A.; Rodgers, A.; Dagg, B.M.; Khatri, B.; Ho, M.M.; Gutierrez, M.G.; Cortes, T.; de Carvalho, L.P.S. Comparative fitness analysis of D-cycloserine resistant mutants reveals both fitness-neutral and high-fitness cost genotypes. Nat. Commun. 2019, 10, 4177. [Google Scholar] [CrossRef] [Green Version]
- Almeida, D.; Ioerger, T.; Tyagi, S.; Li, S.Y.; Mdluli, K.; Andries, K.; Grosset, J.; Sacchettini, J.; Nuermberger, E. Mutations in pepQ Confer Low-Level Resistance to Bedaquiline and Clofazimine in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2016, 60, 4590–4599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Chen, J.; Cui, P.; Shi, W.; Zhang, W.; Zhang, Y. Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2015, 70, 2507–2510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pi, R.; Liu, Q.; Jiang, Q.; Gao, Q. Characterization of linezolid-resistance-associated mutations in Mycobacterium tuberculosis through WGS. J. Antimicrob. Chemother. 2019, 74, 1795–1798. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Ismail, N.A.; Omar, S.V.; Peters, R.P.H. In Vitro Study of Stepwise Acquisition of rv0678 and atpE Mutations Conferring Bedaquiline Resistance. Antimicrob. Agents Chemother. 2019, 63, e00292-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, N.; Omar, S.V.; Ismail, N.A.; Peters, R.P.H. Collated data of mutation frequencies and associated genetic variants of bedaquiline, clofazimine and linezolid resistance in Mycobacterium tuberculosis. Data Brief 2018, 20, 1975–1983. [Google Scholar] [CrossRef]
- Shi, W.; Chen, J.; Zhang, S.; Zhang, W.; Zhang, Y. Identification of Novel Mutations in LprG (rv1411c), rv0521, rv3630, rv0010c, ppsC, and cyp128 Associated with Pyrazinoic Acid/Pyrazinamide Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2018, 62, e00430-18. [Google Scholar] [CrossRef] [Green Version]
- Jackson, N.; Czaplewski, L.; Piddock, L.J.V. Discovery and development of new antibacterial drugs: Learning from experience? J. Antimicrob. Chemother. 2018, 73, 1452–1459. [Google Scholar] [CrossRef] [Green Version]
- Venugopala, K.N.; Chandrashekharappa, S.; Deb, P.K.; Tratrat, C.; Pillay, M.; Chopra, D.; Al-Shar’i, N.A.; Hourani, W.; Dahabiyeh, L.A.; Borah, P.; et al. Anti-tubercular activity and molecular docking studies of indolizine derivatives targeting mycobacterial InhA enzyme. J. Enzym. Inhib. Med. Chem. 2021, 36, 1472–1487. [Google Scholar] [CrossRef]
- Campos, A.I.; Zampieri, M. Metabolomics-Driven Exploration of the Chemical Drug Space to Predict Combination Antimicrobial Therapies. Mol. Cell 2019, 74, 1291–1303.e6. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.C.S.; Nelson, J.; VanderSluis, B.; Deshpande, R.; Butts, A.; Kagan, S.; Polacheck, I.; Krysan, D.J.; Myers, C.L.; Madhani, H.D. Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell 2014, 159, 1168–1187. [Google Scholar] [CrossRef] [Green Version]
- Wambaugh, M.A.; Shakya, V.P.S.; Lewis, A.J.; Mulvey, M.A.; Brown, J.C.S. High-throughput identification and rational design of synergistic small-molecule pairs for combating and bypassing antibiotic resistance. PLoS Biol. 2017, 15, e2001644. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Sieron, A.; King, A.M.; De Pascale, G.; Pawlowski, A.C.; Koteva, K.; Wright, G.D. A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chem. Biol. 2017, 24, 98–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namdari, P.; Negahdari, B.; Eatemadi, A. Synthesis, properties and biomedical applications of carbon-based quantum dots: An updated review. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 87, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Kotta, S.; Aldawsari, H.M.; Badr-Eldin, S.M.; Alhakamy, N.A.; Md, S.; Nair, A.B.; Deb, P.K. Exploring the Potential of Carbon Dots to Combat COVID-19. Front. Mol. Biosci. 2020, 7, 616575. [Google Scholar] [CrossRef] [PubMed]
- Knoblauch, R.; Harvey, A.; Ra, E.; Greenberg, K.M.; Lau, J.; Hawkins, E.; Geddes, C.D. Antimicrobial carbon nanodots: Photodynamic inactivation and dark antimicrobial effects on bacteria by brominated carbon nanodots. Nanoscale 2021, 13, 85–99. [Google Scholar] [CrossRef]
- Yuan, F.; Li, S.; Fan, Z.; Meng, X.; Fan, L.; Yang, S. Shining carbon dots: Synthesis and biomedical and optoelectronic applications. Nano Today 2016, 11, 565–586. [Google Scholar] [CrossRef]
- Knoblauch, R.; Geddes, C.D. Carbon Nanodots in Photodynamic Antimicrobial Therapy: A Review. Materials 2020, 13, 4004. [Google Scholar] [CrossRef]
- Mandal, S.; Prasad, S.R.; Mandal, D.; Das, P. Bovine Serum Albumin Amplified Reactive Oxygen Species Generation from Anthrarufin-Derived Carbon Dot and Concomitant Nanoassembly for Combination Antibiotic-Photodynamic Therapy Application. ACS Appl. Mater. Interfaces 2019, 11, 33273–33284. [Google Scholar] [CrossRef]
- Knoblauch, R.; Ra, E.; Geddes, C.D. Heavy carbon nanodots 2: Plasmon amplification in Quanta Plate™ wells and the correlation with the synchronous scattering spectrum. Phys. Chem. Chem. Phys. PCCP 2019, 21, 1254–1259. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, X.; Tang, D.; Wu, S.; Hou, X.; Liu, J.; Wu, P. Phosphorescent Carbon Dots for Highly Efficient Oxygen Photosensitization and as Photo-oxidative Nanozymes. ACS Appl. Mater. Interfaces 2018, 10, 40808–40814. [Google Scholar] [CrossRef]
- Pham, S.H.; Choi, Y.; Choi, J. Stimuli-Responsive Nanomaterials for Application in Antitumor Therapy and Drug Delivery. Pharmaceutics 2020, 12, 630. [Google Scholar] [CrossRef] [PubMed]
- Devnarain, N.; Osman, N.; Fasiku, V.O.; Makhathini, S.; Salih, M.; Ibrahim, U.H.; Govender, T. Intrinsic stimuli-responsive nanocarriers for smart drug delivery of antibacterial agents-An in-depth review of the last two decades. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnology 2021, 13, e1664. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Rasheed, T.; Nabeel, F.; Hayat, U.; Bilal, M.; Iqbal, H.M.N. Endogenous and Exogenous Stimuli-Responsive Drug Delivery Systems for Programmed Site-Specific Release. Molecules 2019, 24, 1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.Y.; Gao, J.; Wang, Z. Bioresponsive Nanoparticles Targeted to Infectious Microenvironments for Sepsis Management. Adv. Mater. (Deerfield Beach Fla.) 2018, 30, e1803618. [Google Scholar] [CrossRef]
- Moradi, S.; Barati, A.; Salehi, E.; Tonelli, A.E.; Hamedi, H. Preparation and characterization of chitosan based hydrogels containing cyclodextrin inclusion compounds or nanoemulsions of thyme oil. Polym. Int. 2019, 68, 1891–1902. [Google Scholar] [CrossRef]
- Mao, C.; Xie, X.; Liu, X.; Cui, Z.; Yang, X.; Yeung, K.W.K.; Pan, H.; Chu, P.K.; Wu, S. The controlled drug release by pH-sensitive molecularly imprinted nanospheres for enhanced antibacterial activity. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 77, 84–91. [Google Scholar] [CrossRef]
- Cao, J.; Zhao, Y.; Liu, Y.; Tian, S.; Zheng, C.; Liu, C.; Zhai, Y.; An, Y.; Busscher, H.J.; Shi, L. Phosphorylcholine-based polymer encapsulated chitosan nanoparticles enhance the penetration of antimicrobials in a staphylococcal biofilm. ACS Macro Lett. 2019, 8, 651–657. [Google Scholar] [CrossRef]
- Niaz, T.; Shabbir, S.; Noor, T.; Abbasi, R.; Imran, M. Alginate-caseinate based pH-responsive nano-coacervates to combat resistant bacterial biofilms in oral cavity. Int. J. Biol. Macromol. 2020, 156, 1366–1380. [Google Scholar] [CrossRef]
- Chahardahmasoumi, S.; Sarvi, M.N.; Jalali, S.A.H. Modified montmorillonite nanosheets as a nanocarrier with smart pH-responsive control on the antimicrobial activity of tetracycline upon release. Appl. Clay Sci. 2019, 178, 105135. [Google Scholar] [CrossRef]
- Lääveri, T.; Sterne, J.; Rombo, L.; Kantele, A. Systematic review of loperamide: No proof of antibiotics being superior to loperamide in treatment of mild/moderate travellers’ diarrhoea. Travel Med. Infect. Dis. 2016, 14, 299–312. [Google Scholar] [CrossRef]
- King, A.M.; Reid-Yu, S.A.; Wang, W.; King, D.T.; De Pascale, G.; Strynadka, N.C.; Walsh, T.R.; Coombes, B.K.; Wright, G.D. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510, 503–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hense, B.A.; Schuster, M. Core principles of bacterial autoinducer systems. Microbiol. Mol. Biol. Rev. MMBR 2015, 79, 153–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Z.; Guo, Z.; Zhou, L.; Wang, Y.; Zhang, J.; Hu, J.; Zhang, Y. Biomembrane induced in situ self-assembly of peptide with enhanced antimicrobial activity. Biomater. Sci. 2020, 8, 2031–2039. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Liu, Y.; Huang, X.; Ding, S.; Wang, Y.; Shen, J.; Zhu, K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020, 5, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E. New antibiotics and antimicrobial combination therapy for the treatment of gram-negative bacterial infections. Curr. Opin. Crit. Care 2015, 21, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Antonoplis, A.; Zang, X.; Huttner, M.A.; Chong, K.K.L.; Lee, Y.B.; Co, J.Y.; Amieva, M.R.; Kline, K.A.; Wender, P.A.; Cegelski, L. A Dual-Function Antibiotic-Transporter Conjugate Exhibits Superior Activity in Sterilizing MRSA Biofilms and Killing Persister Cells. J. Am. Chem. Soc. 2018, 140, 16140–16151. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Neundorf, I. Design and Application of Antimicrobial Peptide Conjugates. Int. J. Mol. Sci. 2016, 17, 701. [Google Scholar] [CrossRef] [Green Version]
- Tetali, S.R.; Kunapaeddi, E.; Mailavaram, R.P.; Singh, V.; Borah, P.; Deb, P.K.; Venugopala, K.N.; Hourani, W.; Tekade, R.K. Current advances in the clinical development of anti-tubercular agents. Tuberculosis 2020, 125, 101989. [Google Scholar] [CrossRef]
- Wright, G.D. Environmental and clinical antibiotic resistomes, same only different. Curr. Opin. Microbiol. 2019, 51, 57–63. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 26 January 2022).
- Bloom, G.; Merrett, G.B.; Wilkinson, A.; Lin, V.; Paulin, S. Antimicrobial resistance and universal health coverage. BMJ Glob. Health 2017, 2, e000518. [Google Scholar] [CrossRef]
- Pokharel, S.; Raut, S.; Adhikari, B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob. Health 2019, 4, e002104. [Google Scholar] [CrossRef] [PubMed]












| Era | Year(s) | Approach |
|---|---|---|
| Golden era | 1940–1962 | Research-based on natural products Identification based on Whole-Cell screening |
| The era of Medicinal chemistry | 1950–1980 | Synthetic tweaking Identification based on Whole-Cell screening Compounds showing broad-spectrum activity |
| The era of antimicrobial Resistance | 1960 onwards | Modern methods of drug discovery Target-based drug design Ligand-based drug design Compounds showing broad-spectrum activity |
| Era of narrow-spectrum | 2025 | Unconventional methods for drug design and discovery Combinatorial synthesis Diagnostic development |
| Database | Description |
|---|---|
| Collection of antimicrobial peptides (CAMP) | Holding experimentally validated and predicted AMP sequences |
| AMPer | Database and automated discovery tool that is used for gene-coded AMPs |
| Antimicrobial Peptide Database (APD) | Containing the AMPs from natural sources (~98%) |
| BACTIBASE | Data repository of bacteriocin AMPs |
| PhytAMP | Database of plant base antimicrobial peptides |
| RAPD | Database of AMPs produced by recombinant technology |
| HIPdb | Peptides showing anti-HIV activity |
| Bagel2 | A tool for bacteriocin mining |
| Peptaibol | Database for peptaibols |
| PenBase | Database for penaeidins |
| Defensins Knowledge Base | Database for defensins |
| CyBase | Database for cyclotides |
| Drug Group | Drug | Target Gene | Reference |
|---|---|---|---|
| A | Levofloxacin (14) or Moxifluxacin (15) | gyrA | [114] |
| Bedaquiline (16) | atpE | [115,116] | |
| Linezolid (17) | rplC | [117] | |
| B | Clofazimine (18) | rv0678 rv1979c | [118] |
| Cycloserine (19) or Terizidone (20) | Rv2535c alr | [119,120] | |
| C | Ethambutol (21) | embCAB | [121,122] |
| Delamanid (22) | ddn fgd-1 fbiA fbiB fbiC | [123] | |
| Pyrazinamide (23) | pncA rpsA panD clpc1 | [124] | |
| Imipenem (24)/Clavulanic acid (35) or Meropenem (25) | rv2518c rv3682 rv2068c | [125] | |
| Amikacin (26) | Rrs | [126] | |
| Streptomycin (27) | Psl | [127] | |
| Ethionamide (28) or Prothionamide (29) | Rv0565c ethA mymA katG inhA | [128] | |
| Paraminosalicylic acid (30) | thyA folC dfrA | [129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shinu, P.; Mouslem, A.K.A.; Nair, A.B.; Venugopala, K.N.; Attimarad, M.; Singh, V.A.; Nagaraja, S.; Alotaibi, G.; Deb, P.K. Progress Report: Antimicrobial Drug Discovery in the Resistance Era. Pharmaceuticals 2022, 15, 413. https://doi.org/10.3390/ph15040413
Shinu P, Mouslem AKA, Nair AB, Venugopala KN, Attimarad M, Singh VA, Nagaraja S, Alotaibi G, Deb PK. Progress Report: Antimicrobial Drug Discovery in the Resistance Era. Pharmaceuticals. 2022; 15(4):413. https://doi.org/10.3390/ph15040413
Chicago/Turabian StyleShinu, Pottathil, Abdulaziz K. Al Mouslem, Anroop B. Nair, Katharigatta N. Venugopala, Mahesh Attimarad, Varsha A. Singh, Sreeharsha Nagaraja, Ghallab Alotaibi, and Pran Kishore Deb. 2022. "Progress Report: Antimicrobial Drug Discovery in the Resistance Era" Pharmaceuticals 15, no. 4: 413. https://doi.org/10.3390/ph15040413
APA StyleShinu, P., Mouslem, A. K. A., Nair, A. B., Venugopala, K. N., Attimarad, M., Singh, V. A., Nagaraja, S., Alotaibi, G., & Deb, P. K. (2022). Progress Report: Antimicrobial Drug Discovery in the Resistance Era. Pharmaceuticals, 15(4), 413. https://doi.org/10.3390/ph15040413