Promising Antibacterial and Antifungal Agents Based on Thiolated Vitamin K3 Analogs: Synthesis, Bioevaluation, Molecular Docking
Abstract
:1. Introduction
2. Results and Discussion
2.1. Library Design and Synthesis
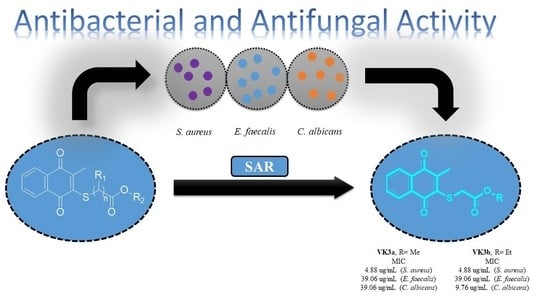
2.2. Biological Activity
2.2.1. Evaluation of Antimicrobial Activity of the Thiolated VK3 Analogs (VK3a–g)
2.2.2. Toxicity Evaluation
2.2.3. Hemolytic Activity
2.2.4. Time-Kill Kinetic Study
2.2.5. Evaluation of the In Vitro Antibiofilm Activity
2.2.6. Molecular Docking Study
2.2.7. In Silico Drug-Likeness and ADME Analysis of Selected Thiolated VK3 Analogs
3. Materials and Methods
3.1. Chemicals and Apparatus
3.2. X-ray Diffraction Analysis
3.3. General Procedure for the Synthesis of the Thiolated Vitamin K3 (VK3) Analogs
3.3.1. Methyl 2-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)acetate (VK3a)
3.3.2. Ethyl 2-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)acetate (VK3b)
3.3.3. Ethyl 2-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)propanoate(VK3c)
3.3.4. Methyl 3-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)propanoate (VK3d)
3.3.5. Ethyl 3-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)propanoate (VK3e)
3.3.6. Butyl 3-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)propanoate (VK3f)
3.3.7. 2-Ethylhexyl 2-(3-Methyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylthio)acetate (VK3g)
3.4. Biological Evaluation
3.4.1. MIC Evaluation
3.4.2. Determination of Time-Kill Curves
3.4.3. Determination of the Antibiofilm Activities
3.4.4. Hemolysis Assay
3.5. Statistical Analysis
3.6. Molecular Docking
3.7. In Silico Drug-Likeness and ADMET Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fairfield, K.M.; Fletcher, R.H. Vitamins for chronic disease prevention in adults: Scientific review. JAMA 2002, 287, 3116–3126. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, H.; Khiali, S.; Rezaei, H.; Rezaee, H.; Bannazadeh Baghi, H.; Pourghasem, M.; Entezari-Maleki, T. Potential Roles of Vitamins in the Management of COVID-19: A Comprehensive Review. Pharm. Sci. 2021, 27, S29–S49. [Google Scholar] [CrossRef]
- Jovic, T.H.; Ali, S.R.; Ibrahim, N.; Jessop, Z.M.; Tarassoli, S.P.; Dobbs, T.D.; Holford, P.; Thornton, C.A.; Whitaker, I.S. Could Vitamins Help in the Fight Against COVID-19? Nutrients 2020, 12, 2550. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, G.; Chen, D.; Lu, Y. β-Carotene and astaxanthin with human and bovine serum albumins. Food Chem. 2015, 179, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Bourassa, P.; Hasni, I.; Tajmir-Riahi, H.A. Folic acid complexes with human and bovine serum albumins. Food Chem. 2011, 129, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, Q.; Zuo, Y.; Bi, Y.; Gao, S. Spectroscopic Studies on the Interaction of Vitamin C with Bovine Serum Albumin. J. Solut. Chem. 2009, 38, 15–25. [Google Scholar] [CrossRef]
- Haddad, J.G. Human serum binding protein for vitamin D and its metabolites (DBP): Evidence that actin is the DBP binding component in human skeletal muscle. Arch. Biochem. Biophys. 1982, 213, 538–544. [Google Scholar] [CrossRef]
- Dahlbäck, B. Purification of human vitamin K-dependent protein S and its limited proteolysis by thrombin. Biochem. J. 1983, 209, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Suganthi, M.; Elango, K.P. Synthesis, characterization and serum albumin binding studies of vitamin K3 derivatives. J. Photochem. Photobiol. B 2017, 166, 126–135. [Google Scholar] [CrossRef]
- Hamidi, M.S.; Cheung, A.M. Vitamin K and musculoskeletal health in postmenopausal women. Mol. Nutr. Food Res. 2014, 58, 1647–1657. [Google Scholar] [CrossRef]
- Scheiber, D.; Veulemans, V.; Horn, P.; Chatrou, M.L.; Potthoff, S.A.; Kelm, M.; Schurgers, L.J.; Westenfeld, R. High-Dose Menaquinone-7 Supplementation Reduces Cardiovascular Calcification in a Murine Model of Extraosseous Calcification. Nutrients 2015, 7, 6991–7011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yerramsetti, N.; Dampanaboina, L.; Mendu, V.; Battula, S. Synergistic factors ensue high expediency in the synthesis of menaquinone [K-2] analogue MK-6: Application to access an efficient one-pot protocol to MK-9. Tetrahedron 2020, 76, 131696. [Google Scholar] [CrossRef]
- Li, X.Y.; Himes, R.A.; Prosser, L.C.; Christie, C.F.; Watt, E.; Edwards, S.F.; Metcalf, C.S.; West, P.J.; Wilcox, K.S.; Chan, S.S.L.; et al. Discovery of the First Vitamin K Analogue as a Potential Treatment of Pharmacoresistant Seizures. J. Med. Chem. 2020, 63, 5865–5878. [Google Scholar] [CrossRef] [PubMed]
- Wellington, K.W.; Hlatshwayo, V.; Kolesnikova, N.I.; Saha, S.T.; Kaur, M.; Motadi, L.R. Anticancer activities of vitamin K3 analogues. Investig. New Drug 2020, 38, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, J.H. Effect of Anticancer Quinones on Reactive Oxygen Production by Adult Rat Heart Myocytes. Oxidative Med. Cell. Longev. 2020, 2020, 8877100. [Google Scholar] [CrossRef]
- Bauer, H.; Fritz-Wolf, K.; Winzer, A.; Kuhner, S.; Little, S.; Yardley, V.; Vezin, H.; Palfey, B.; Schirmer, R.H.; Davioud-Charvet, E. A fluoro analogue of the menadione derivative 6-[2′-(3′-methyl)-1′,4′-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme. J. Am. Chem. Soc. 2006, 128, 10784–10794. [Google Scholar] [CrossRef] [PubMed]
- Patel, O.P.S.; Beteck, R.M.; Legoabe, L.J. Antimalarial application of quinones: A recent update. Eur. J. Med. Chem. 2021, 210, 113084. [Google Scholar] [CrossRef]
- Majiene, D.; Kuseliauskyte, J.; Stimbirys, A.; Jekabsone, A. Comparison of the Effect of Native 1,4-Naphthoquinones Plumbagin, Menadione, and Lawsone on Viability, Redox Status, and Mitochondrial Functions of C6 Glioblastoma Cells. Nutrients 2019, 11, 1294. [Google Scholar] [CrossRef] [Green Version]
- Deniz, N.G.; Abdassalam, A.F.S.; Ozyurek, M.; Yesil, E.A.; Sayil, C. New vitamin K3 (menadione) analogues: Synthesis, characterization, antioxidant and catalase inhibition activities. J. Chem. Sci. 2020, 132, 138. [Google Scholar] [CrossRef]
- Hileman, E.O.; Liu, J.S.; Albitar, M.; Keating, M.J.; Huang, P. Intrinsic oxidative stress in cancer cells: A biochemical basis for therapeutic selectivity. Cancer Chemoth. Pharm. 2004, 53, 209–219. [Google Scholar] [CrossRef]
- Tetef, M.; Margolin, K.; Ahn, C.; Akman, S.; Chow, W.; Leong, L.; Morgan, R.J., Jr.; Raschko, J.; Somlo, G.; Doroshow, J.H. Mitomycin C and menadione for the treatment of lung cancer: A phase II trial. Investig. New Drugs 1995, 13, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Osada, S.; Tomita, H.; Tanaka, Y.; Tokuyama, Y.; Tanaka, H.; Sakashita, F.; Takahashi, T. The utility of vitamin K3 (menadione) against pancreatic cancer. Anticancer Res. 2008, 28, 45–50. [Google Scholar] [PubMed]
- Verrax, J.; Cadrobbi, J.; Marques, C.; Taper, H.; Habraken, Y.; Piette, J.; Calderon, P.B. Ascorbate potentiates the cytotoxicity of menadione leading to an oxidative stress that kills cancer cells by a non-apoptotic caspase-3 independent form of cell death. Apoptosis 2004, 9, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S.; Raghu, D.; Karunagaran, D. Menadione (Vitamin K3) Induces Apoptosis of Human Oral Cancer Cells and Reduces their Metastatic Potential by Modulating the Expression of Epithelial to Mesenchymal Transition Markers and Inhibiting Migration. Asian Pac. J. Cancer P 2013, 14, 5461–5465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, E.O.; Sun, T.P.; Chang, J.Y.; Wang, C.C.; Chi, K.H.; Cheng, A.L.; Nutter, L.M. Menadione-Induced DNA Damage in a Human Tumor-Cell Line. Biochem. Pharmacol. 1991, 42, 1961–1968. [Google Scholar] [CrossRef]
- Zhang, Z.; Ding, Y.; Li, J.; Wang, L.; Xin, X.; Yan, J.; Wu, J.; Yuan, A.; Hu, Y. Versatile iron-vitamin K3 derivative-based nanoscale coordination polymer augments tumor ferroptotic therapy. Nano Res. 2021, 14, 2398–2409. [Google Scholar] [CrossRef]
- Bayrak, N.; Ciftci, H.I.; Yildiz, M.; Yildirim, H.; Sever, B.; Tateishi, H.; Otsuka, M.; Fujita, M.; Tuyun, A.F. Structure based design, synthesis, and evaluation of anti-CML activity of the quinolinequinones as LY83583 analogs. Chem. Biol. Interact. 2021, 345, 109555. [Google Scholar] [CrossRef]
- Mataraci-Kara, E.; Bayrak, N.; Yildirim, H.; Yildiz, M.; Ataman, M.; Ozbek-Celik, B.; Tuyun, A.F. Plastoquinone analogs: A potential antimicrobial lead structure intensely suppressing Staphylococcus epidermidis and Candida albicans growth. Med. Chem. Res. 2021, 30, 1728–1737. [Google Scholar] [CrossRef]
- Jannuzzi, A.T.; Yıldız, M.; Bayrak, N.; Yıldırım, H.; Shilkar, D.; Jayaprakash, V.; TuYuN, A.F. Anticancer agents based on Plastoquinone analogs with N-phenylpiperazine: Structure-activity relationship and mechanism of action in breast cancer cells. Chem.-Biol. Interact. 2021, 349, 109673. [Google Scholar] [CrossRef]
- Yildirim, H. Arylthio analogs of 5,8-quinolinedione: Synthesis, characterization, antibacterial, and antifungal evaluations. Phosphorus Sulfur 2020, 195, 516–522. [Google Scholar] [CrossRef]
- Yildirim, H. Synthesis, characterization, and biological evaluation of a set of new alkylthio substituted plastoquinones containing ester group. J. Mol. Struct. 2020, 1203, 127433. [Google Scholar] [CrossRef]
- Ibis, C.; Tuyun, A.F.; Bahar, H.; Ayla, S.S.; Stasevych, M.V.; Musyanovych, R.Y.; Komarovska-Porokhnyavets, O.; Novikov, V. Synthesis of novel 1,4-naphthoquinone derivatives: Antibacterial and antifungal agents. Med. Chem. Res. 2013, 22, 2879–2888. [Google Scholar] [CrossRef]
- Ibis, C.; Tuyun, A.F.; Bahar, H.; Ayla, S.S.; Stasevych, M.V.; Musyanovych, R.Y.; Komarovska-Porokhnyavets, O.; Novikov, V. Nucleophilic substitution reactions of 1,4-naphthoquinone and biologic properties of novel S-, S,S-, N-, and N,S-substituted 1,4-naphthoquinone derivatives. Med. Chem. Res. 2014, 23, 2140–2149. [Google Scholar] [CrossRef]
- Espinosa-Bustos, C.; Vázquez, K.; Varela, J.; Cerecetto, H.; Paulino, M.; Segura, R.; Pizarro, J.; Vera, B.; González, M.; Zarate, A.M.; et al. New aryloxy-quinone derivatives with promising activity on Trypanosoma cruzi. Arch. Pharm. 2020, 353, 1900213. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, P.D.O.; Jobim, G.D.B.; Ferreira, C.C.D.; Bernardes, L.R.; Dias, R.B.; Sales, C.B.S.; Valverde, L.D.; Rocha, C.A.G.; Soares, M.B.P.; Bezerra, D.P.; et al. A new synthetic antitumor naphthoquinone induces ROS-mediated apoptosis with activation of the JNK and p38 signaling pathways. Chem.-Biol. Interact. 2021, 343, 109444. [Google Scholar] [CrossRef]
- Mataraci-Kara, E.; Bayrak, N.; Yildiz, M.; Yildirim, H.; TuYu, N.A. Active Quinolinequinones against Methicillin-Resistant Staphylococcus spp. Chem. Biodivers 2021, 19, e202100616. [Google Scholar] [CrossRef]
- Ciftci, H.I.; Bayrak, N.; Yıldız, M.; Yıldırım, H.; Sever, B.; Tateishi, H.; Otsuka, M.; Fujita, M.; Tuyun, A.F. Design, synthesis and investigation of the mechanism of action underlying anti-leukemic effects of the quinolinequinones as LY83583 analogs. Bioorg. Chem. 2021, 114, 105160. [Google Scholar] [CrossRef]
- Bayrak, N.; Yildiz, M.; Yildirim, H.; Mataraci-Kara, E.; Tuyun, A.F. Novel plastoquinone analogs containing benzocaine and its analogs: Structure-based design, synthesis, and structural characterization. Res. Chem. Intermediat. 2021, 47, 2125–2141. [Google Scholar] [CrossRef]
- Bayrak, N.; Yildiz, M.; Yildirim, H.; Kara, E.M.; Celik, B.O.; Tuyun, A.F. Brominated plastoquinone analogs: Synthesis, structural characterization, and biological evaluation. J. Mol. Struct. 2020, 1219, 128560. [Google Scholar] [CrossRef]
- Wellington, K.W.; Kolesnikova, N.I.; Nyoka, N.B.P.; McGaw, L.J. Investigation of the antimicrobial and anticancer activity of aminonaphthoquinones. Drug Dev. Res. 2019, 80, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Wellington, K.W. Understanding cancer and the anticancer activities of naphthoquinones—A review. Rsc. Adv. 2015, 5, 20309–20338. [Google Scholar] [CrossRef]
- Ryu, C.K.; Song, A.L.; Lee, J.Y.; Hong, J.A.; Yoon, J.H.; Kim, A. Synthesis and antifungal activity of benzofuran-5-ols. Bioorg. Med. Chem. Lett. 2010, 20, 6777–6780. [Google Scholar] [CrossRef] [PubMed]
- Ibis, C.; Ozsoy-Gunes, Z.; Tuyun, A.F.; Ayla, S.S.; Bahar, H.; Stasevych, M.V.; Musyanovych, R.Y.; Komarovska-Porokhnyavets, O.; Novikov, V. Synthesis, antibacterial and antifungal evaluation of thio- or piperazinyl-substituted 1,4-naphthoquinone derivatives. J. Sulfur. Chem. 2016, 37, 477–487. [Google Scholar] [CrossRef]
- Ravichandiran, P.; Maslyk, M.; Sheet, S.; Janeczko, M.; Premnath, D.; Kim, A.R.; Park, B.H.; Han, M.K.; Yoo, D.J. Synthesis and Antimicrobial Evaluation of 1,4-Naphthoquinone Derivatives as Potential Antibacterial Agents. Chemistryopen 2019, 8, 589–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravichandiran, P.; Sheet, S.; Premnath, D.; Kim, A.R.; Yoo, D.J. 1,4-Naphthoquinone Analogues: Potent Antibacterial Agents and Mode of Action Evaluation. Molecules 2019, 24, 1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravichandiran, P.; Subramaniyan, S.A.; Kim, S.Y.; Kim, J.S.; Park, B.H.; Shim, K.S.; Yoo, D.J. Synthesis and Anticancer Evaluation of 1,4-Naphthoquinone Derivatives Containing a Phenylaminosulfanyl Moiety. Chemmedchem 2019, 14, 532–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tandon, V.K.; Maurya, H.K.; Tripathi, A.; ShivaKeshava, G.B.; Shukla, P.K.; Srivastava, P.; Panda, D. 2,3-Disubstituted-1,4-naphthoquinones, 12H-benzo[b]phenothiazine-6, 11-diones and related compounds: Synthesis and Biological evaluation as potential antiproliferative and antifungal agents. Eur. J. Med. Chem. 2009, 44, 1086–1092. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; p. 19087. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1997. [Google Scholar]
- Ricci, M.S.; Zong, W.-X. Chemotherapeutic Approaches for Targeting Cell Death Pathways. Oncologist 2006, 11, 342–357. [Google Scholar] [CrossRef] [Green Version]
- Boyd, M.R.; Pauli, K.D. Some Practical Considerations and Applications of the National-Cancer-Institute in-Vitro Anticancer Drug Discovery Screen. Drug Develop. Res. 1995, 34, 91–109. [Google Scholar] [CrossRef]
- Razaghi, A.; Heimann, K.; Schaeffer, P.M.; Gibson, S.B. Negative regulators of cell death pathways in cancer: Perspective on biomarkers and targeted therapies. Apoptosis 2018, 23, 93–112. [Google Scholar] [CrossRef]
- Da Silva, R.E.; Ribeiro, F.d.O.S.; de Carvalho, A.M.A.; Daboit, T.C.; Marinho-Filho, J.D.B.; Matos, T.S.; Pessoa, O.D.L.; de Souza de Almeida Leite, J.R.; de Araújo, A.R.; dos Santos Soares, M.J. Antimicrobial and antibiofilm activity of the benzoquinone oncocalyxone A. Microb. Pathogenesis. 2020, 149, 104513. [Google Scholar] [CrossRef] [PubMed]
- Novais, J.S.; Carvalho, M.F.; Ramundo, M.S.; Beltrame, C.O.; Geraldo, R.B.; Jordao, A.K.; Ferreira, V.F.; Castro, H.C.; Figueiredo, A.M.S. Antibiofilm effects of N,O-acetals derived from 2-amino-1,4-naphthoquinone are associated with downregulation of important global virulence regulators in methicillin-resistant Staphylococcus aureus. Sci. Rep 2020, 10, 19631. [Google Scholar] [CrossRef] [PubMed]
- Mataraci-Kara, E.; Ozbek-Celik, B. Investigation of the effects of various antibiotics against Klebsiella pneumoniae biofilms on in vitro catheter model. J. Chemotherapy 2018, 30, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, M.; Le Senechal, C.; Brözel, V.S.; Costaglioli, P.; Barthe, C.; Bonneu, M.; Garbay, B.; Vilain, S. Exploring early steps in biofilm formation: Set-up of an experimental system for molecular studies. BMC Microbiol. 2014, 14, 253. [Google Scholar] [CrossRef] [Green Version]
- Petit, C.M.; Koretke, K.K. Characterization of Streptococcus pneumoniae thymidylate kinase: Steady-state kinetics of the forward reaction and isothermal titration calorimetry. Biochem. J. 2002, 363, 825–831. [Google Scholar] [CrossRef]
- Keating, T.A.; Newman, J.V.; Olivier, N.B.; Otterson, L.G.; Andrews, B.; Boriack-Sjodin, P.A.; Breen, J.N.; Doig, P.; Dumas, J.; Gangl, E.; et al. In Vivo Validation of Thymidylate Kinase (TMK) with a Rationally Designed, Selective Antibacterial Compound. Acs Chem. Biol. 2012, 7, 1866–1872. [Google Scholar] [CrossRef]
- Kawatkar, S.P.; Keating, T.A.; Olivier, N.B.; Breen, J.N.; Green, O.M.; Guler, S.Y.; Hentemann, M.F.; Loch, J.T.; McKenzie, A.R.; Newman, J.V.; et al. Antibacterial Inhibitors of Gram-Positive Thymidylate Kinase: Structure–Activity Relationships and Chiral Preference of a New Hydrophobic Binding Region. J. Med. Chem. 2014, 57, 4584–4597. [Google Scholar] [CrossRef]
- Vanheusden, V.; Munier-Lehmann, H.; Froeyen, M.; Busson, R.; Rozenski, J.; Herdewijn, P.; Van Calenbergh, S. Discovery of Bicyclic Thymidine Analogues as Selective and High-Affinity Inhibitors of Mycobacterium tuberculosis Thymidine Monophosphate Kinase. J. Med. Chem. 2004, 47, 6187–6194. [Google Scholar] [CrossRef] [Green Version]
- Abo-Salem, H.M.; Abd El Salam, H.A.; Abdel-Aziem, A.; Abdel-Aziz, M.S.; El-Sawy, E.R. Synthesis, Molecular Docking, and Biofilm Formation Inhibitory Activity of Bis(Indolyl)Pyridines Analogues of the Marine Alkaloid Nortopsentin. Molecules 2021, 26, 4112. [Google Scholar] [CrossRef]
- Dechouk, L.F.; Bouchoucha, A.; Abdi, Y.; Si Larbi, K.; Bouzaheur, A.; Terrachet-Bouaziz, S. Coordination of new palladium (II) complexes with derived furopyran-3,4-dione ligands: Synthesis, characterization, redox behaviour, DFT, antimicrobial activity, molecular docking and ADMET studies. J. Mol. Struct. 2022, 1257, 132611. [Google Scholar] [CrossRef]
- Martínez-Botella, G.; Loch, J.T.; Green, O.M.; Kawatkar, S.P.; Olivier, N.B.; Boriack-Sjodin, P.A.; Keating, T.A. Sulfonylpiperidines as novel, antibacterial inhibitors of Gram-positive thymidylate kinase (TMK). Bioorg. Med. Chem. Lett. 2013, 23, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Teague, S.J.; Davis, A.M.; Leeson, P.D.; Oprea, T. The Design of Leadlike Combinatorial Libraries. Angew. Chem. Int. Ed. Engl. 1999, 38, 3743–3748. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M., Jr.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Bruker. APEX2, Version 2014.1-1; Bruker AXS Inc.: Madison, WI, USA, 2014. [Google Scholar]
- Bruker. SAINT, Version 8.34A; Bruker AXS Inc.: Madison, WI, USA, 2013. [Google Scholar]
- Bruker. SADABS, Version 2012/2; Bruker AXS Inc.: Madison, WI, USA, 2012. [Google Scholar]
- Bruker. SHELXTL, Version 6.14; Bruker AXS Inc.: Madison, WI, USA, 2000. [Google Scholar]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. D 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van De Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Habib, N.S.; Mahran, M.A. Synthesis and biological evaluation of novel naphthoquinone derivatives as potential anticancer and antimicrobial agents. Boll. Chim. Farm. 2004, 143, 299–307. [Google Scholar]
- Nagar, B.; Dhar, B.B. Visible Light-Mediated Thiolation of Substituted 1,4-Naphthoquinones Using Eosin Y as a Photoredox Catalyst. J. Org. Chem. 2022, 87, 3195–3201. [Google Scholar] [CrossRef] [PubMed]
- National Committee for Clinical Laboratory Standards. Methods for Determining Bactericidal Activity of Antimicrobial Agents; NCCLS: Wayne, MI, USA, 1999. [Google Scholar]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides. In Antimicrobial Peptides: Methods and Protocols; Hansen, P.R., Ed.; Springer New York: New York, NY, USA, 2017; pp. 427–435. [Google Scholar]
- Tateishi, H.; Tateishi, M.; Radwan, M.O.; Masunaga, T.; Kawatashiro, K.; Oba, Y.; Oyama, M.; Inoue-Kitahashi, N.; Fujita, M.; Okamoto, Y.; et al. A New Inhibitor of ADAM17 Composed of a Zinc-Binding Dithiol Moiety and a Specificity Pocket-Binding Appendage. Chem. Pharm. Bull. 2021, 69, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Mingle, D.; Ospanov, M.; Radwan, M.O.; Ashpole, N.; Otsuka, M.; Ross, S.A.; Walker, L.A.; Shilabin, A.G.; Ibrahim, M.A. First in class (S,E)-11-[2-(arylmethylene)hydrazono]-PBD analogs as selective CB2 modulators targeting neurodegenerative disorders. Med. Chem. Res. 2021, 30, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]








| Identification Code | VK3a |
|---|---|
| Chemical formula | C14H12O4S |
| Formula weight (g mol−1) | 276.30 |
| Temperature (K) | 273 |
| Radiation λ (Å) | 0.71073 |
| Crystal system | Monoclinic |
| Space groups, Z | P 1 21/n 1, 4 |
| Unit cell dimensions (Å) | a = 17.1194(16) |
| b = 3.9184(4) | |
| c = 19.3277(18) | |
| α, γ = 90° β = 101.4320(10)° | |
| Volume (Å3) | 1270.8(2) |
| Crystal sizes (mm) | 0.087 × 0.143 × 0.360 |
| dcalc (g cm−3) | 1.444 |
| Absorption coefficient (mm−1) | 0.261 |
| Absorption correction, Tmin, Tmax | none, 0.9780 and 0.9120 |
| θmax, deg | 1.77 to 27.48 |
| Goodness-of-fit on F2 | 1.018 |
| Index ranges | −22 ≤ h ≤ 22 |
| −5 ≤ k ≤ 5 | |
| −25 ≤ l ≤ 25 | |
| Reflections collected | 15,493 |
| Independent reflections | 2896 [R(int) = 0.0558] |
| Final R indices [I > 2σ(I)] | R1 = 0.0417 |
| wR2 = 0.1064 | |
| R indices (all data) | R1 = 0.0587 |
| wR2 = 0.1162 | |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 2896/0/174 |
| Largest diff. peak and hole (eÅ−3) | 0.279 and −0.231 |
| S1-C4 | 1.7583(18) | S1-C3 | 1.8145(18) |
| O2-C2 | 1.325(2) | O2-C1 | 1.449(2) |
| O3-C14 | 1.218(2) | O1-C2 | 1.195(2) |
| O4-C7 | 1.216(2) | C2-C3 | 1.500(3) |
| C4-S1-C3 | 102.70(8) | C2-O2-C1 | 116.90(16) |
| C5-C4-C14 | 121.34(16) | C5-C4-S1 | 119.68(13) |
| C14-C4-S1 | 118.61(12) | C12-C13-C8 | 119.37(17) |
| C4-C5-C6 | 123.16(17) | C7-C5-C6 | 116.23(15) |
| O1-C2-O2 | 124.19(18) | O1-C2-C3 | 122.32(18) |
| O2-C2-C3 | 113.48(16) | C2-C3-S1 | 112.18(12) |
| C3-S1-C4-C5 | 137.42(16) | C3-S1-C4-C14 | −49.49(16) |
| C1-O2-C2-O1 | 0.2(3) | C8-C13-C14-O3 | −169.01(19) |
| C6-C5-C7-O4 | 0.8(3) | S1-C4-C14-O3 | −5.4(3) |
| C9-C8-C7-O4 | −1.6(3) | C1-O2-C2-C3 | −178.98(18) |
| O1-C2-C3-S1 | 164.08(18) | O2-C2-C3-S1 | −16.7(2) |
| C4-S1-C3-C2 | −151.94(14) | C4-C5-C7-O4 | 179.9(2) |
| .D-H···A | D-H | H···A | D···A | D-H···A |
| C3-H3A···O3 i | 0.97 | 2.56 | 3.513 (2) | 168 |
| Thiolated VK3 | Substituents | Gram-Negative Bacteria a (MIC, μg/mL) | Gram-Positive Bacteria b (MIC, μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| General Formula | ID | R1 | R2 | Pa | Ec | Kp | Pm | Sa | Se | Ef |
 | VK3a (n = 1) | H | -CH3 | - | - | - | - | 4.88 | - | 39.06 |
| VK3b (n = 1) | H | -CH2CH3 | - | - | - | - | 4.88 | - | 39.06 | |
| VK3c (n = 1) | CH3 | -CH2CH3 | - | - | - | - | 9.76 | - | 39.06 | |
| VK3d (n = 2) | H | -CH3 | - | - | - | - | 4.88 | - | 39.06 | |
| VK3e (n = 2) | H | -CH2CH3 | - | - | - | - | 312.50 | - | - | |
| VK3f (n = 2) | H | -(CH2)3CH3 | - | - | - | - | 9.76 | - | 312.50 | |
| VK3g (n = 1) | H | -CH2CH(n-C2H5)(CH2)3CH3 | - | - | - | - | 312.50 | - | - | |
| Ceftazidime | 2.44 | |||||||||
| Cefuroxime-Na | 4.88 | 4.88 | 2.44 | 1.22 | ||||||
| Cefuroxime | 9.76 | |||||||||
| Amikacin | 128.00 | |||||||||
| Thiolated VK3 | Substituents | Fungi a (MIC, μg/mL) | ||||
|---|---|---|---|---|---|---|
| General Formula | ID | R1 | R2 | Ca | Cp | Ct |
 | VK3a (n = 1) | H | -CH3 | 39.06 | 78.12 | 9.76 |
| VK3b (n = 1) | H | -CH2CH3 | 9.76 | 78.12 | 19.53 | |
| VK3c (n = 1) | CH3 | -CH2CH3 | 39.06 | 78.12 | 39.06 | |
| VK3d (n = 2) | H | -CH3 | 39.06 | 156.25 | 9.76 | |
| VK3e (n = 2) | H | -CH2CH3 | - | - | - | |
| VK3f (n = 2) | H | -(CH2)3CH3 | 78.12 | - | 19.53 | |
| VK3g (n = 1) | H | -CH2CH(n-C2H5)(CH2)3CH3 | - | - | 625 | |
| Clotrimazole | 4.9 | |||||
| Amphotericin B | 0.5 | 1 | ||||
| Molecules | Number of Strains MIC (MBC) | MIC Range | MIC50 | MIC90 | MBC Range | MBC50 | MBC90 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentrations (µg/mL) | ||||||||||||||||
| >625 | 625 | 312.50 | 156.25 | 78.12 | 39.06 | 19.53 | 9.76 | 4.88 | 2.44 | |||||||
| VK3a | - (1) | 4 (3) | - (1) | 1 (4) | 1 (3) | 10 (6) | 2 (1) | 2 (1) | 9.76–625 | 39.06 | 625 | 9.76–>625 | 78.12 | 625 | ||
| VK3b | - (1) | - (1) | 1 (4) | 1 (2) | 6 (4) | 8 (6) | 4 (2) | 2.44–39.06 | 4.88 | 9.76 | 2.44–156.25 | 9.76 | 39.06 | |||
| Property/Rule | VK3a | VK3b | |
|---|---|---|---|
| Physico-chemical properties and drug-likeness prediction | MW | 276 | 290 |
| Log P | 2.34 | 2.62 | |
| TPSA Å2 | 85.74 | 85.74 | |
| HBA | 4 | 4 | |
| HBD | 0 | 0 | |
| Lipiniski | Yes, 0 violation | Yes, 0 violation | |
| Ghose | Yes | Yes | |
| Veber | Yes | Yes | |
| Egan | Yes | Yes | |
| Muegge | Yes | Yes | |
| Leadlikeness | Yes | Yes | |
| ADME Prediction | BBB permeability | No | No |
| GI absorption | High | high | |
| Log S | −3.08 | −3.31 | |
| Solubility | Soluble | Soluble | |
| Bioavailability score | 0.55 | 0.55 | |
| CYP1A2, CYP2C19, CYP2C9 | Yes | Yes | |
| CYP2D6, CYP3A4 | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yıldırım, H.; Yıldız, M.; Bayrak, N.; Mataracı-Kara, E.; Radwan, M.O.; Jannuzzi, A.T.; Otsuka, M.; Fujita, M.; TuYuN, A.F. Promising Antibacterial and Antifungal Agents Based on Thiolated Vitamin K3 Analogs: Synthesis, Bioevaluation, Molecular Docking. Pharmaceuticals 2022, 15, 586. https://doi.org/10.3390/ph15050586
Yıldırım H, Yıldız M, Bayrak N, Mataracı-Kara E, Radwan MO, Jannuzzi AT, Otsuka M, Fujita M, TuYuN AF. Promising Antibacterial and Antifungal Agents Based on Thiolated Vitamin K3 Analogs: Synthesis, Bioevaluation, Molecular Docking. Pharmaceuticals. 2022; 15(5):586. https://doi.org/10.3390/ph15050586
Chicago/Turabian StyleYıldırım, Hatice, Mahmut Yıldız, Nilüfer Bayrak, Emel Mataracı-Kara, Mohamed Osman Radwan, Ayse Tarbin Jannuzzi, Masami Otsuka, Mikako Fujita, and Amaç Fatih TuYuN. 2022. "Promising Antibacterial and Antifungal Agents Based on Thiolated Vitamin K3 Analogs: Synthesis, Bioevaluation, Molecular Docking" Pharmaceuticals 15, no. 5: 586. https://doi.org/10.3390/ph15050586
APA StyleYıldırım, H., Yıldız, M., Bayrak, N., Mataracı-Kara, E., Radwan, M. O., Jannuzzi, A. T., Otsuka, M., Fujita, M., & TuYuN, A. F. (2022). Promising Antibacterial and Antifungal Agents Based on Thiolated Vitamin K3 Analogs: Synthesis, Bioevaluation, Molecular Docking. Pharmaceuticals, 15(5), 586. https://doi.org/10.3390/ph15050586