A Synopsis of Hepatitis C Virus Treatments and Future Perspectives
Abstract
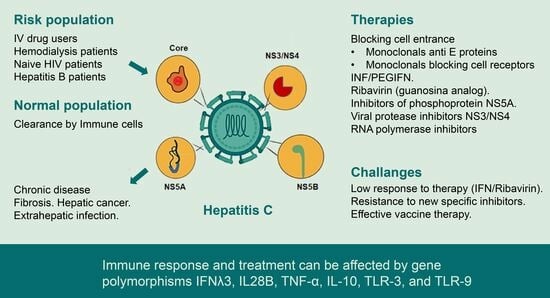
:1. Introduction
1.1. Viral Genome
1.2. Viral Transmission and Viral Receptors
1.3. Antiviral Response and Immune Response Activation
1.4. Coinfections: HBV–HCV and HCV–HIV
2. Interferon α and Ribavirin
3. Direct-Acting Antivirals (DAA)
3.1. NS3/4 Protease Inhibitors
3.1.1. Telaprevir and Boceprevir
3.1.2. Asunaprevir
3.1.3. Simeprevir
3.1.4. Paritaprevir
3.1.5. Grazoprevir
3.1.6. Glecaprevir and Voxileprevir
3.2. NS5B Polymerase Inhibitors
3.2.1. Nucleotide: Sofosbuvir
3.2.2. Non-Nucleotide: Dasabuvir
3.3. NS5A Polymerase Inhibitors
3.3.1. Ledipasvir (LDV)
3.3.2. Daclatasvir (DCV)
3.3.3. Ombitasvir
3.3.4. Elbasvir
3.3.5. Velpatasvir and Pibrentasvir
4. Treatment in Coinfection (HCV/HIV, HBV/HCV, HBV/HCV/HIV)
5. Treatment of Pregnant Women, Vertical Transmission and Paediatric Care
6. Host Genetics, Infection and Response to HCV Treatments
7. Resistance-Associated Substitutions (RAS)
8. Other Strategies
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ghany, M.G.; Lok, A.S.F.; Dienstag, J.L.; Feinstone, S.M.; Hoofnagle, J.H.; Jake Liang, T.; Seeff, L.B.; Cohen, D.E.; Bezerra, J.A.; Chung, R.T. The 2020 Nobel Prize for Medicine or Physiology for the Discovery of Hepatitis C Virus: A Triumph of Curiosity and Persistence. Hepatology 2021, 74, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Kao, J.H. Acute hepatitis C virus infection: Clinical update and remaining challenges. Clin. Mol. Hepatol. 2023, 29, 623–642. [Google Scholar] [CrossRef]
- Dubuisson, J.; Cosset, F.L. Virology and cell biology of the hepatitis C virus life cycle—An update. J. Hepatol. 2014, 61, S3–S13. [Google Scholar] [CrossRef]
- Lee, M.H.; Yang, H.I.; Yuan, Y.; L’Italien, G.; Chen, C.J. Epidemiology and natural history of hepatitis C virus infection. World J. Gastroenterol. 2014, 20, 9270–9280. [Google Scholar] [PubMed]
- Alazard-Dany, N.; Denolly, S.; Boson, B.; Cosset, F.L. Overview of HCV Life Cycle with a Special Focus on Current and Possible Future Antiviral Targets. Viruses 2019, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Basit, H.; Tyagi, I.; Koirala, J. Hepatitis C; StatPearls Publishing: Treasure Island, FL, USA, 2023; Available online: https://www.ncbi.nlm.nih.gov/books/NBK430897/ (accessed on 28 September 2023).
- Hofmeister, M.G.; Rosenthal, E.M.; Barker, L.K.; Rosenberg, E.S.; Barranco, M.A.; Hall, E.W.; Edlin, B.R.; Mermin, J.; Ward, J.W.; Ryerson, A.B. Estimating Prevalence of Hepatitis C Virus Infection in the United States, 2013–2016. Hepatology 2019, 69, 1020–1031. [Google Scholar] [CrossRef]
- Stasi, C.; Silvestri, C.; Voller, F. Update on Hepatitis C Epidemiology: Unaware and Untreated Infected Population Could Be the Key to Elimination. SN Compr. Clin. Med. 2020, 2, 2808–2815. [Google Scholar] [CrossRef]
- Fénéant, L.; Levy, S.; Cocquerel, L. CD81 and hepatitis C virus (HCV) infection. Viruses 2014, 6, 535–572. [Google Scholar] [CrossRef]
- Westhaus, S.; Deest, M.; Nguyen, A.T.; Stanke, F.; Heckl, D.; Costa, R.; Schambach, A.; Manns, M.P.; Berg, T.; Vondran, F.W.; et al. Scavenger receptor class B member 1 (SCARB1) variants modulate hepatitis C virus replication cycle and viral load. J. Hepatol. 2017, 67, 237–245. [Google Scholar] [CrossRef]
- Liu, S.; Yang, W.; Shen, L.; Turner, J.R.; Coyne, C.B.; Wang, T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 2009, 83, 2011–2014. [Google Scholar] [CrossRef]
- Lupberger, J.; Zeisel, M.B.; Xiao, F.; Thumann, C.; Fofana, I.; Zona, L.; Davis, C.; Mee, C.J.; Turek, M.; Gorke, S.; et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011, 17, 589–595. [Google Scholar] [CrossRef]
- Mazumdar, B.; Banerjee, A.; Meyer, K.; Ray, R. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology 2011, 54, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Lavie, M.; Dubuisson, J. Interplay between hepatitis C virus and lipid metabolism during virus entry and assembly. Biochimie 2017, 141, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Ray, K. NPC1L1 identified as a novel HCV entry factor. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Martinez, P.; Séron, K.; Luo, G.; Allain, F.; Dubuisson, J.; Belouzard, S. Characterization of hepatitis C virus interaction with heparan sulfate proteoglycans. J. Virol. 2015, 89, 3846–3858. [Google Scholar] [CrossRef]
- Marino, R.; Deibis, L.; De Sanctis, J.B.; Bianco, N.E.; Toro, F. Interaction of immune complexes isolated from hepatitis C virus-infected individuals with human cell lines. Med. Microbiol. Immunol. 2005, 194, 73–80. [Google Scholar] [CrossRef]
- Matsumori, A.; Shimada, M.; Obata, T. Leukocytes are the major target of hepatitis C virus infection: Possible mechanism of multiorgan involvement including the heart. CVD Prev. Control 2010, 5, 51–58. [Google Scholar] [CrossRef]
- Toro, F.; Conesa, A.; Garcia, A.; Deibis, L.; Bianco, N.E.; De Sanctis, J.B. HCV RNA sequences in eosinophils of chronic HCV-infected patients. J. Med. 1999, 30, 279–288. [Google Scholar]
- Corado, J.; Toro, F.; Rivera, H.; Bianco, N.E.; Deibis, L.; De Sanctis, J.B. Impairment of natural killer (NK) cytotoxic activity in hepatitis C virus (HCV) infection. Clin. Exp. Immunol. 1997, 109, 451–457. [Google Scholar] [CrossRef]
- Marciniewicz, E.; Podgórski, P.; Pawłowski, T.; Małyszczak, K.; Fleischer-Stępniewska, K.; Knysz, B.; Waliszewska-Prosół, M.; Żelwetro, A.; Rymer, W.; Inglot, M.; et al. Evaluation of brain volume alterations in HCV-infected patients after interferon-free therapy: A pilot study. J. Neurol. Sci. 2019, 399, 36–43. [Google Scholar] [CrossRef]
- Waliszewska-Prosół, M.; Bladowska, J.; Ejma, M.; Fleischer-Stępniewska, K.; Rymer, W.; Sąsiadek, M.; Pawłowski, T.; Małyszczak, K.; Inglot, M.; Żelwetro, A.; et al. Visual and brainstem auditory evoked potentials in HCV-infected patients before and after interferon-free therapy—A pilot study. Int. J. Infect. Dis. 2019, 80, 122–128. [Google Scholar] [CrossRef]
- Dustin, L.B.; Bartolini, B.; Capobianchi, M.R.; Pistello, M. Hepatitis C virus: Life cycle in cells, infection and host response, and analysis of molecular markers influencing the outcome of infection and response to therapy. Clin. Microbiol. Infect. 2016, 22, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Janiak, M.; Cortes, K.C.; Demkow, U.; Radkowski, M. Spontaneous Elimination of Hepatitis C Virus Infection. Adv. Exp. Med. Biol. 2018, 1039, 45–54. [Google Scholar]
- Chigbu, D.I.; Loonawat, R.; Sehgal, M.; Patel, D.; Jain, P. Hepatitis C Virus Infection: Host−Virus Interaction and Mechanisms of Viral Persistence. Cells 2019, 8, 376. [Google Scholar] [CrossRef]
- Axley, P.; Ahmed, Z.; Ravi, S.; Singal, A.K. Hepatitis C Virus and Hepatocellular Carcinoma: A Narrative Review. J. Clin. Transl. Hepatol. 2018, 6, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Feld, J.J.; Hoofnagle, J.H. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 2005, 436, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Kim, S.S.; Yeung, E.; Kamegaya, Y.; Blackard, J.T.; Kim, K.A.; Holtzman, M.J.; Chung, R.T. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J. Virol. 2006, 80, 9226–9235. [Google Scholar] [CrossRef]
- Zhao, L.J.; He, S.F.; Liu, Y.; Zhao, P.; Bian, Z.Q.; Qi, Z.T. Inhibition of STAT Pathway Impairs Anti-Hepatitis C Virus Effect of Interferon Alpha. Cell. Physiol. Biochem. 2016, 40, 77–90. [Google Scholar] [CrossRef]
- Yamauchi, S.; Takeuchi, K.; Chihara, K.; Honjoh, C.; Kato, Y.; Yoshiki, H.; Hotta, H.; Sada, K. STAT1 is essential for the inhibition of hepatitis C virus replication by interferon-λ but not by interferon-α. Sci. Rep. 2016, 6, 38336. [Google Scholar] [CrossRef]
- Polyak, S.J.; Khabar, K.S.; Rezeiq, M.; Gretch, D.R. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J. Virol. 2007, 5, 6209–6211. [Google Scholar] [CrossRef]
- Taylor, D.R.; Shi, S.T.; Romano, P.R.; Barber, G.N.; Lai, M.M. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 1999, 285, 107–110. [Google Scholar] [CrossRef]
- Cesaro, T.; Michiels, T. Inhibition of PKR by Viruses. Front. Micobiol. 2021, 12, 757238. [Google Scholar] [CrossRef]
- Wang, H.; Xin, X.; Wang, M.; Han, L.; Li, J.; Hao, Y.; Zheng, C.; Shen, C. Myxovirus resistance protein A inhibits hepatitis C virus replication through JAK-STAT pathway activation. Arch. Virol. 2018, 163, 1429–1438. [Google Scholar] [CrossRef]
- Barbaglia, M.N.; Harris, J.M.; Smirnov, A.; Burlone, M.E.; Rigamonti, C.; Pirisi, M.; Minisini, R.; Magri, A. 17β-Oestradiol Protects from Hepatitis C Virus Infection through Induction of Type I Interferon. Viruses 2022, 14, 1806. [Google Scholar] [CrossRef] [PubMed]
- Marchi, E.; Ramamurthy, N.; Ansari, M.A.; E Harrer, C.; Barnes, E.; Klenerman, P. Defining the key intrahepatic gene networks in HCV infection driven by sex. Gut 2023, 72, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.D.; Salinas, E.; Grakoui, A. Immune system control of hepatitis C virus infection. Curr. Opin. Virol. 2021, 46, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Dustin, L.B. Innate and Adaptive Immune Responses in Chronic HCV Infection. Curr. Drug Targets 2017, 18, 826–843. [Google Scholar] [CrossRef]
- Corado, J.A.; Toro, F.I.; Baroja, M.L.; Bianco, N.E.; Machado, I.V. CD3- and CD28-activating pathways in HCV infection. Viral. Immunol. 1994, 7, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Auma, A.W.N.; Shive, C.L.; Kostadinova, L.; Anthony, D.D. Variable Normalisation of Naïve CD4+ Lymphopenia and Markers of Monocyte and T Cell Activation over the Course of Direct-Acting Antiviral Treatment of Chronic Hepatitis C Virus Infection. Viruses 2021, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Ferrufino, R.Q.; Rodrigues, C.; Figueiredo, G.M.; Gleison, D.; Yapura, S.; de Matos, M.L.M.; Witkin, S.S.; Mendes-Correa, M.C. Factors Associated with Spontaneous Clearance of Recently Acquired Hepatitis C Virus among HIV-Positive Men in Brazil. Viruses 2023, 15, 314. [Google Scholar] [CrossRef]
- Shin, E.C.; Sung, P.; Park, S.H. Immune responses and immunopathology in acute and chronic viral hepatitis. Nat. Rev. Immunol. 2016, 16, 509–523. [Google Scholar] [CrossRef]
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014, 59, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Marascio, N.; Torti, C.; Liberto, M.C.; Focá, A. Update on different aspects of HCV variability: Focus on NS5B polymerase. BMC Infect. Dis. 2014, 14 (Suppl. S5), S1. [Google Scholar] [CrossRef] [PubMed]
- Echeverría, N.; Moratorio, G.; Cristina, J.; Moreno, P. Hepatitis C virus genetic variability and evolution. World J. Hepatol. 2015, 7, 831–845. [Google Scholar] [CrossRef]
- Gower, E.; Estes, C.; Blach, S.; Razavi-Shearer, K.; Razavi, H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol. 2014, 61, S45–S57. [Google Scholar] [CrossRef]
- Petruzziello, A.; Marigliano, S.; Loquercio, G.; Cozzolino, A.; Cacciapuoti, C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J. Gastroenterol. 2016, 22, 7824–7840. [Google Scholar] [CrossRef]
- Gobran, S.T.; Ancuta, P.; Shoukry, N.H. A Tale of Two Viruses: Immunological Insights Into HCV/HIV Coinfection. Front. Immunol. 2021, 12, 726419. [Google Scholar] [CrossRef]
- Shahriar, S.; Araf, Y.; Ahmad, R.; Kattel, P.; Sah, G.S.; Rahaman, T.I.; Sadiea, R.Z.; Sultana, S.; Islam, M.S.; Zheng, C.; et al. Insights Into the Coinfections of Human Immunodeficiency Virus-Hepatitis B Virus, Human Immunodeficiency Virus-Hepatitis C Virus, and Hepatitis B Virus-Hepatitis C Virus: Prevalence, Risk Factors, Pathogenesis, Diagnosis, and Treatment. Front. Microbiol. 2022, 12, 780887. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, D.; Deutsch, M. The spectrum of HBV/HCV coinfection: Epidemiology, clinical characteristics, viral interactions and management. Ann. Gastroenterol. 2015, 28, 221–228. [Google Scholar] [PubMed]
- Wieland, S.; Makowska, Z.; Campana, B.; Calabrese, D.; Dill, M.T.; Chung, J.; Chisari, F.V.; Heim, M.H. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology 2013, 59, 2121–2130. [Google Scholar] [CrossRef]
- Nkongolo, S.; Mahamed, D.; Kuipery, A.; Sanchez Vasquez, J.D.; Kim, S.C.; Mehrotra, A.; Patel, A.; Hu, C.; McGilvray, I.; Feld, J.J.; et al. Longitudinal liver sampling in patients with chronic hepatitis B starting antiviral therapy reveals hepatotoxic CD8+ T cells. J. Clin. Investig. 2023, 133, e158903. [Google Scholar] [CrossRef]
- Perales, C.; Beach, N.M.; Gallego, I.; Soria, M.E.; Quer, J.; Esteban, J.I.; Rice, C.; Domingo, E.; Sheldon, J. Response of Hepatitis C Virus to Long-Term Passage in the Presence of Alpha Interferon: Multiple Mutations and a Common Phenotype. J. Virol. 2013, 87, 7593–7607. [Google Scholar] [CrossRef]
- Rojas, J.M.; Alejo, A.; Martín, V.; Sevilla, N. Viral pathogen-induced mechanisms to antagonise mammalian interferon (IFN) signalling pathway. Cell. Mol. Life Sci. 2021, 78, 1423–1444. [Google Scholar] [CrossRef]
- Mertowska, P.; Smolak, K.; Mertowski, S.; Grywalska, E. Immunomodulatory Role of Interferons in Viral and Bacterial Infections. Int. J. Mol. Sci. 2023, 24, 10115. [Google Scholar] [CrossRef]
- Hoofnagle, J.H.; Mullen, K.D.; Jones, D.B.; Rustgi, V.; Di Bisceglie, A.; Peters, M.; Waggoner, J.G.; Park, Y.; Jones, E.A. Treatment of Chronic Non-A, Non-B Hepatitis with Recombinant Human Alpha Interferon. N. Engl. J. Med. 1986, 315, 1575–1578. [Google Scholar] [CrossRef] [PubMed]
- Jaeckel, E.; Mayer, J.; Trautwein, C. Treatment of Acute Hepatitis C with Interferon Alfa-2b. N. Engl. J. Med. 2001, 345, 1452–1457. [Google Scholar] [CrossRef]
- Aghemo, A.; Rumi, M.G.; Colombo, M. Pegylated interferons alpha2a and alpha2b in the treatment of chronic hepatitis C. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 485–494. [Google Scholar] [CrossRef]
- McHutchison, J.G.; Gordon, S.C.; Schiff, E.R.; Shiffman, M.L.; Lee, W.M.; Rustgi, V.K.; Goodman, Z.D.; Ling, M.H.; Cort, S.; Albrecht, J.K. Interferon Alfa-2b Alone or in Combination with Ribavirin as Initial Treatment for Chronic Hepatitis C. N. Engl. J. Med. 1998, 339, 1485–1492. [Google Scholar] [CrossRef] [PubMed]
- Poynard, T.; Marcellin, P.; Lee, S.S.; Niederau, C.; Minuk, G.S.; Ideo, G.; Bain, V.; Heathcote, J.; Zeuzem, S.; Trepo, C.; et al. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet 1998, 352, 1426–1432. [Google Scholar] [CrossRef]
- Thomas, E.; Ghany, M.G.; Liang, T.J. The application and mechanism of action of ribavirin in therapy of hepatitis C. Antivir. Chem. Chemother. 2012, 23, 1–12. [Google Scholar] [CrossRef]
- Fried, M.W.; Shiffman, M.L.; Reddy, K.R.; Smith, C.; Marinos, G.; Gonçales, F.L.; Häussinger, D.; Diago, M.; Carosi, G.; Dhumeaux, D.; et al. Peginterferon Alfa-2a plus Ribavirin for Chronic Hepatitis C Virus Infection. N. Engl. J. Med. 2002, 347, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Hadziyannis, S.J.; Sette, H., Jr.; Morgan, T.R.; Balan, V.; Diago, M.; Marcellin, P.; Ramadori, G.; Bodenheimer, H., Jr.; Bernstein, D.; Rizzetto, M.; et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: A randomised study of treatment duration and ribavirin dose. Ann. Intern. Med. 2004, 140, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Ascione, A.; De Luca, M.; Tartaglione, M.T.; Lampasi, F.; Di Costanzo, G.G.; Lanza, A.G.; Picciotto, F.P.; Marino–Marsilia, G.; Fontanella, L.; Leandro, G. Peginterferon Alfa-2a Plus Ribavirin Is More Effective Than Peginterferon Alfa-2b Plus Ribavirin for Treating Chronic Hepatitis C Virus Infection. Gastroenterology 2010, 138, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Rumi, M.G.; Aghemo, A.; Prati, G.M.; D’Ambrosio, R.; Donato, M.F.; Soffredini, R.; Del Ninno, E.; Russo, A.; Colombo, M. Randomised Study of Peginterferon-α2a Plus Ribavirin vs Peginterferon-α2b Plus Ribavirin in Chronic Hepatitis C. Gastroenterology 2010, 138, 108–115. [Google Scholar] [CrossRef]
- Alavian, S.M.; Behnava, B.; Tabatabaei, S.V. The comparative efficacy and safety of peginterferon alpha-2a vs. 2b for the treatment of chronic HCV infection: A meta-analysis. Hepat. Mon. 2010, 10, 121–131. [Google Scholar]
- Nakamura, M.; Kanda, T.; Miyamura, T.; Wu, S.; Nakamoto, S.; Yokosuka, O. Alanine aminotransferase elevation during peginterferon alpha-2a or alpha-2b plus ribavirin treatment. Int. J. Med. Sci. 2013, 10, 1015–1021. [Google Scholar] [CrossRef]
- Manns, M.P.; McHutchison, J.G.; Gordon, S.C.; Rustgi, V.K.; Shiffman, M.; Reindollar, R.; Goodman, Z.D.; Koury, K.; Ling, M.; Albrecht, J.K. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 2001, 358, 958–965. [Google Scholar] [CrossRef]
- Hu, J.H.; Chang, M.L.; Huang, T.J.; Yeh, C.T.; Chiu, W.N.; Chiang, M.S.; Chen, M.Y. Comparison of Compliance and Efficacy of Pegylated Interferon α-2a and α-2b in Adults with Chronic Hepatitis C. J. Interf. Cytokine Res. 2019, 39, 205–213. [Google Scholar] [CrossRef]
- Afdhal, N.H.; Dieterich, D.T.; Pockros, P.J.; Schiff, E.R.; Shiffman, M.L.; Sulkowski, M.S.; Wright, T.; Younossi, Z.; Goon, B.L.; Tang, K.L.; et al. Proactive Study Group. Epoetin alfa maintains ribavirin dose in HCV-infected patients: A prospective, double-blind, randomised controlled study. Gastroenterology 2004, 126, 1302–1311. [Google Scholar] [CrossRef]
- Van Thiel, D.H.; Faruki, H.; Friedlander, L.; Fagiuoli, S.; Caraceni, P.; Molloy, P.J.; Kania, R.J.; Wright, H.I. Combination treatment of advanced HCV associated liver disease with interferon and G-CSF. Hepatogastroenterology 1995, 42, 907–912. [Google Scholar]
- Afdhal, N.H.; Dusheiko, G.M.; Giannini, E.G.; Chen, P.; Han, K.; Mohsin, A.; Rodriguez–Torres, M.; Rugina, S.; Bakulin, I.; Lawitz, E.; et al. Eltrombopag increases platelet numbers in thrombocytopenic patients with HCV infection and cirrhosis, allowing for effective antiviral therapy. Gastroenterology 2014, 146, 442–452.e1. [Google Scholar] [CrossRef]
- O’Leary, J.G.; Davis, G.L. Hepatitis C virus replication and potential targets for direct-acting agents. Ther. Adv. Gastroenterol. 2010, 3, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Milani, A.; Basimi, P.; Agi, E.; Bolhassani, A. Pharmaceutical Approaches for Treatment of Hepatitis C virus. Curr. Pharm. Des. 2020, 26, 4304–4314. [Google Scholar] [CrossRef] [PubMed]
- Bernal, L.A.; Soti, V. Hepatitis C Virus: Insights Into Its History, Treatment, Challenges, and Future Directions. Cureus 2023, 15, e43924. [Google Scholar] [CrossRef]
- Failla, C.; Tomei, L.; De Francesco, R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 1994, 68, 3753–3760. [Google Scholar] [CrossRef]
- Tabata, K.; Neufeldt, J.C.; Bartenschlager, R. Hepatitis C Virus Replication. Cold Spring Harb. Perspect. Med. 2020, 10, a037093. [Google Scholar] [CrossRef]
- Kiang, T.K.; Wilby, K.J.; Ensom, M.H. Telaprevir: Clinical pharmacokinetics, pharmacodynamics, and drug-drug interactions. Clin. Pharmacokinet. 2013, 52, 487–510. [Google Scholar] [CrossRef]
- Chang, M.H.; Gordon, L.A.; Fung, H.B. Boceprevir: A protease inhibitor for the treatment of hepatitis C. Clin. Ther. 2012, 34, 2021–2038. [Google Scholar] [CrossRef]
- Gordon, S.C.; Reddy, K.R.; Jacobson, I.M.; Poordad, F.; Bronowicki, J.P.; Bacon, B.; Buti, M.; Hu, K.Q.; Pedicone, L.D.; Burroughs, M.; et al. Boceprevir plus peginterferon α-2b/ribavirin in chronic hepatitis C genotype 1: Impact of baseline viral load on sustained virologic response. J. Clin. Gastroenterol. 2014, 48, 435–443. [Google Scholar] [CrossRef]
- Akamatsu, N.; Sugawara, Y.; Kokudo, N. Asunaprevir (BMS-650032) for the treatment of hepatitis C virus. Expert Rev. Anti-Infect. Ther. 2015, 13, 1307–1317. [Google Scholar] [CrossRef]
- Osawa, M.; Ueno, T.; Shiozaki, T.; Li, H.; Garimella, T. Safety Exposure-Response Analysis for Daclatasvir, Asunaprevir, and Beclabuvir Combinations in HCV-Infected Subjects. J. Clin. Pharmacol. 2019, 59, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Bao, Y.; Xia, W.; Wu, H.; Wei, F.; Zhang, Y.; Zhang, R.; Xu, X. Resistance-associated mutations to HCV protease inhibitors naturally pre-existed in HIV/HCV coinfected, treatment-naïve patients. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Childs-Kean, L.M.; Hand, E.O. Simeprevir and sofosbuvir for treatment of chronic hepatitis C infection. Clin. Ther. 2015, 37, 243–267. [Google Scholar] [CrossRef]
- Sulkowski, M.S.; Vargas, H.E.; Di Bisceglie, A.M.; Kuo, A.; Reddy, K.R.; Lim, J.K.; Morelli, G.; Darling, J.M.; Feld, J.J.; Brown, R.S.; et al. Effectiveness of Simeprevir Plus Sofosbuvir, with or without Ribavirin, in Real-World Patients with HCV Genotype 1 Infection. Gastroenterology 2016, 150, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Klibanov, O.M.; Gale, S.E.; Santevecchi, B. Ombitasvir/paritaprevir/ritonavir and dasabuvir tablets for hepatitis C virus genotype 1 infection. Ann. Pharmacother. 2015, 49, 566–581. [Google Scholar] [CrossRef]
- Deeks, E.D. Ombitasvir/Paritaprevir/Ritonavir Plus Dasabuvir: A Review in Chronic HCV Genotype 1 Infection. Drugs 2015, 75, 1027–1038. [Google Scholar] [CrossRef]
- Al-Salama, Z.T.; Deeks, E.D. Elbasvir/Grazoprevir: A Review in Chronic HCV Genotypes 1 and 4. Drugs 2017, 77, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Abushouk, A.I.; Menshawy, A.; Attia, A.; Mohamed, A.; Negida, A.; Abdel-Daim, M. Meta-Analysis of Grazoprevir plus Elbasvir for Treatment of Hepatitis C Virus Genotype 1 Infection. Ann. Hepatol. 2018, 17, 18–32. [Google Scholar] [CrossRef]
- Lamb, Y.N. Glecaprevir/Pibrentasvir: First Global Approval. Drugs 2017, 77, 1797–1804. [Google Scholar] [CrossRef]
- Chahine, E.B.; Kelley, D.; Childs-Kean, L.M. Sofosbuvir/Velpatasvir/Voxilaprevir: A Pan-Genotypic Direct-Acting Antiviral Combination for Hepatitis C. Ann. Pharmacother. 2018, 52, 352–363. [Google Scholar] [CrossRef]
- Puoti, M.; Panzeri, C.; Rossotti, R.; Baiguera, C. Efficacy of sofosbuvir-based therapies in HIV/HCV infected patients and persons who inject drugs. Dig. Liver Dis. 2014, 46 (Suppl. S5), S206–S211. [Google Scholar] [CrossRef]
- Smith, M.A.; Chan, J.; Mohammad, R.A. Ledipasvir-sofosbuvir: Interferon-/ribavirin-free regimen for chronic hepatitis C virus infection. Ann. Pharmacother. 2015, 49, 343–350. [Google Scholar] [CrossRef]
- Chen, L.; Du, L.; Kang, S.; Ma, F.; Li, C.; He, M.; Bai, L.; Tang, H. Sofosbuvir plus Ribavirin is effective for HCV elimination in people living with HIV from rural area of China. Sci. Rep. 2021, 11, 11301. [Google Scholar] [CrossRef]
- Gentile, I.; Buonomo, A.R.; Borgia, G. Dasabuvir: A Non-Nucleoside Inhibitor of NS5B for the Treatment of Hepatitis C Virus Infection. Rev. Recent Clin. Trials 2014, 9, 115–123. [Google Scholar] [CrossRef]
- Gamal, N.; Andreone, P. Working together to tackle HCV infection: Ombitasvir/paritaprevir/ritonavir and dasabuvir combination. Drugs Today 2015, 51, 303–314. [Google Scholar] [CrossRef] [PubMed]
- King, J.R.; Zha, J.; Khatri, A.; Dutta, S.; Menon, R.M. Clinical Pharmacokinetics of Dasabuvir. Clin. Pharmacokinet. 2017, 56, 1115–1124. [Google Scholar] [CrossRef]
- Gitto, S.; Gamal, N.; Andreone, P. NS5A inhibitors for the treatment of hepatitis C infection. J. Viral. Hepat. 2017, 24, 180–186. [Google Scholar] [CrossRef]
- Bourlière, M.; Adhoute, X.; Ansaldi, C.; Oules, V.; Benali, S.; Portal, I.; Castellani, P.; Halfon, P. Sofosbuvir plus ledipasvir in combination for the treatment of hepatitis C infection. Expert Rev. Gastroent. Hepatol. 2015, 9, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Belperio, P.S.; Shahoumian, T.A.; Loomis, T.P.; Mole, L.A.; Backus, L.I. Real-world effectiveness of daclatasvir plus sofosbuvir and velpatasvir/sofosbuvir in hepatitis C genotype 2 and 3. J. Hepatol. 2019, 70, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Sabry, N.; Kamel, A.M.; Cordie, A.; Esmat, G. Daclatasvir as a hepatitis C infection treatment option: An up-to-date evaluation. Expert Opin. Pharmacother. 2023, 24, 159–170. [Google Scholar] [CrossRef]
- Lim, S.G.; Aghemo, A.; Chen, P.-J.; Dan, Y.Y.; Gane, E.; Gani, R.; Gish, R.G.; Guan, R.; Jia, J.D.; Lim, K.; et al. Management of hepatitis C virus infection in the Asia-Pacific region: An update. Lancet. Gastroenterol. Hepatol. 2017, 2, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, Y.; Eley, T.; Fura, A.; Li, W.; Bertz, R.J.; Garimella, T. Daclatasvir: A Review of Preclinical and Clinical Pharmacokinetics. Clin. Pharmacokinet. 2018, 57, 911–928. [Google Scholar] [CrossRef] [PubMed]
- Badri, P.S.; Shuster, D.L.; Dutta, S.; Menon, R.M. Clinical Pharmacokinetics of Ombitasvir. Clin. Pharmacokinet. 2017, 56, 1103–1113. [Google Scholar] [CrossRef]
- Wang, S.J.; Huang, C.F.; Yu, M.L. Elbasvir and grazoprevir for the treatment of hepatitis C. Expert Rev. Anti-Infect. Ther. 2021, 19, 1071–1081. [Google Scholar] [CrossRef]
- Lahser, F.C.; Bystol, K.; Curry, S.; McMonagle, P.; Xia, E.; Ingravallo, P.; Chase, R.; Liu, R.; Black, T.; Hazuda, D.; et al. The Combination of Grazoprevir, a Hepatitis C Virus (HCV) NS3/4A Protease Inhibitor, and Elbasvir, an HCV NS5A Inhibitor, Demonstrates a High Genetic Barrier to Resistance in HCV Genotype 1a Replicons. Antimicrob. Agents Chemother. 2016, 60, 2954–2964. [Google Scholar] [CrossRef]
- Ren, X.D.; Fu, X.; He, Y.Q.; Li, C.Y.; Guo, M.; Qiao, M. Safety and efficacy of sofosbuvir-velpatasvir: A meta-analysis. Medicine 2022, 101, e31183. [Google Scholar] [CrossRef]
- Ghany, M.G. AASLD-IDSA Hepatitis C Guidance Panel. Hepatitis C Guidance 2019 Update: American Association for the Study of Liver Diseases-Infectious Diseases Society of America Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Hepatology 2020, 71, 686–721. [Google Scholar] [CrossRef]
- Pearlman, B.; Perrys, M.; Hinds, A. Sofosbuvir/Velpatasvir/Voxilaprevir for Previous Treatment Failures With Glecaprevir/Pibrentasvir in Chronic Hepatitis C Infection. Am. J. Gastroenterol. 2019, 114, 1550–1552. [Google Scholar] [CrossRef]
- Reau, N.; Cheng, W.H.; Shao, Q.; Marx, S.E.; Brooks, H.; Martinez, A. Real-World Effectiveness of 8-Week Glecaprevir/Pibrentasvir in Treatment-Naïve, Compensated Cirrhotic HCV Patients. Infect. Dis. Ther. 2023, 12, 1849–1860. [Google Scholar] [CrossRef]
- Solomon, S.S.; Wagner-Cardoso, S.; Smeaton, L.; Sowah, L.; Wimbish, C.; Robbins, G.; Brates, I.; Scello, C.; Son, A.; Avihingsanon, A.; et al. A minimal monitoring approach for the treatment of hepatitis C virus infection (ACTG A5360 [MINMON]): A phase 4, open-label, single-arm trial. Lancet Gastroenterol. Hepatol. 2022, 7, 307–317. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Aronsohn, A.; Price, J.; Lo Re, V.; AASLD-IDSA HCV Guidance Panel. Hepatitis C Guidance 2023 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin. Infect. Dis. 2023, ciad319. [Google Scholar] [CrossRef]
- Mavilia, M.G.; Wu, G.Y. HBV-HCV Coinfection: Viral Interactions, Management, and Viral Reactivation. J. Clin. Transl. Hepatol. 2018, 6, 296–305. [Google Scholar] [CrossRef]
- Pisaturo, M.; Macera, M.; Alessio, L.; Calò, F.; Coppola, N. Hepatitis B Virus (HBV) Reactivation Following Pharmacological Eradication of Hepatitis C Virus (HCV). Viruses 2019, 11, 850. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, Q.; Sumrin, A.; Iqbal, M.; Younas, S.; Hussain, N.; Mahnoor, M.; Wajid, A. Hepatitis C virus/Hepatitis B virus coinfection: Current prospectives. Antivir. Ther. 2023, 28, 13596535231189643. [Google Scholar] [CrossRef] [PubMed]
- Cairoli, V.; Valle-Millares, D.; Terrón-Orellano, M.C.; Luque, D.; Ryan, P.; Dominguez, L.; Martín-Carbonero, L.; Santos, I.D.L.; De Matteo, E.; Ameigeiras, B.; et al. MicroRNA signature from extracellular vesicles of HCV/HIV coinfected individuals differs from HCV mono-infected. J. Mol. Med. 2023, in press. [Google Scholar] [CrossRef]
- Mirzaei, R.; Karampoor, S.; Korotkova, N.L. The emerging role of miRNA-122 in infectious diseases: Mechanisms and potential biomarkers. Pathol. Res. Pract. 2023, 249, 154725. [Google Scholar] [CrossRef] [PubMed]
- Benova, L.; Mohamoud, Y.A.; Calvert, C.; Abu-Raddad, L.J. Vertical Transmission of Hepatitis C Virus: Systematic Review and Meta-analysis. Clin. Infect. Dis. 2014, 59, 765–773. [Google Scholar] [CrossRef]
- US Preventive Services Task Force; Owens, D.K.; Davidson, K.W.; Krist, A.H.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Donahue, K.; Doubeni, C.A.; Epling, J.W.; et al. Screening for Hepatitis C Virus Infection in Adolescents and Adults: US Preventive Services Task Force Recommendation Statement. JAMA 2020, 323, 970. [Google Scholar]
- Schillie, S.; Wester, C.; Osborne, M.; Wesolowski, L.; Ryerson, A.B. CDC Recommendations for Hepatitis C Screening Among Adults—United States, 2020. MMWR Recomm. Rep. 2020, 69, 1–17. [Google Scholar] [CrossRef]
- Chung, R.T.; Ghany, M.G.; Kim, A.Y.; Marks, K.M.; Naggie, S.; AASLD-IDSA HCV Guidance Panel. Hepatitis C Guidance 2018 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin. Infect. Dis. 2018, 67, 1477–1492. [Google Scholar]
- Kushner, T.; Lange, M.; Sperling, R.; Dieterich, D. Treatment of women with hepatitis C diagnosed in pregnancy: A co-located treatment approach. Gastroenterology 2022, 163, 1454–1456.e1. [Google Scholar] [CrossRef] [PubMed]
- AbdAllah, M.; Alboraie, M.; Abdel-Razek, W.; Hassany, M.; Ammar, I.; Kamal, E.; Alalfy, M.; Okasha, A.; El Akel, W.; Shaaban, E.; et al. Pregnancy outcome of anti-HCV direct-acting antivirals: Real-life data from an Egyptian cohort. Liver Int. 2021, 41, 1494–1497. [Google Scholar] [CrossRef]
- Hughes, B.L.; Page, C.M.; Kuller, J.A. Hepatitis C in pregnancy: Screening, treatment, and management. Am. J. Obstet. Gynecol. 2017, 217, B2–B12. [Google Scholar] [CrossRef] [PubMed]
- Chappell, C.A.; Scarsi, K.K.; Kirby, B.J.; Suri, V.; Gaggar, A.; Bogen, D.L.; Macio, I.S.; Meyn, L.A.; Bunge, K.E.; Krans, E.; et al. Ledipasvir plus sofosbuvir in pregnant women with hepatitis C virus infection: A phase 1 pharmacokinetic study. Lancet Microbe 2020, 1, e200–e208. [Google Scholar] [CrossRef]
- Bitnun, A. The management of infants, children, and youth at risk for hepatitis C virus infection. Paediatr. Child Health. 2021, 26, 440–441. [Google Scholar] [CrossRef]
- Schwarz, K.B.; Rosenthal, P.; Murray, K.F.; Honegger, J.R.; Hardikar, W.; Hague, R.; Mittal, N.; Massetto, B.; Brainard, D.M.; Hsueh, C.; et al. Ledipasvir-Sofosbuvir for 12 Weeks in Children 3 to <6 Years Old With Chronic Hepatitis C. Hepatology 2020, 71, 422–430. [Google Scholar] [PubMed]
- Venkatesh, V.; Seetharaman, K.; Anushree, N. Treatment of hepatitis C in children and adolescents: How far have we reached? World J. Pediatr. 2023, 19, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Peralta, R.P.; Wirth, S.; Squires, R.H.; Mutschler, F.; Lang, T.; Pawlowska, M.; Sluzewski, W.; Majda-Stanislawska, E.; Fischler, B.; Balistreri, W.F.; et al. Elbasvir/grazoprevir in children aged 3–18 years with chronic HCV genotype 1 or 4 infection: A pharmacokinetic modeling study. Hepatol. Commun. 2023, 7, e0031. [Google Scholar] [CrossRef]
- Heck, J.E.; Wu, C.K.; Huang, X.; Chew, K.W.; Tong, M.; Federman, N.; Ritz, B.; Arah, O.A.; Li, C.-Y.; Yu, F.; et al. Cohort study of familial viral hepatitis and risks of paediatric cancers. Int. J. Epidemiol. 2022, 51, 448–457. [Google Scholar] [CrossRef]
- Miri, H.H.; Fazeli, P.; Ali-Hassanzadeh, M.; Bemani, P.; Kabelitz, D.; Kalantar, K. Correlation between IL-28 polymorphism and spontaneous clearance in HCV patients: Systematic review and meta-analysis. Arch. Virol. 2021, 166, 2469–2478. [Google Scholar] [CrossRef]
- Ge, D.; Fellay, J.; Thompson, A.J.; Simon, J.S.; Shianna, K.V.; Urban, T.J.; Heinzen, E.L.; Qiu, P.; Bertelsen, A.H.; Muir, A.J.; et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009, 461, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Watanabe, T.; Tanaka, Y. Role of IL28B for chronic hepatitis C treatment toward personalised medicine. J. Gastroenterol. Hepatol. 2014, 29, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Hayes, C.N.; Imamura, M.; Aikata, H.; Chayama, K. Genetics of IL28B and HCV response to infection and treatment. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 406–417. [Google Scholar] [CrossRef]
- Bhushan, A.; Ghosh, S.; Bhattacharjee, S.; Chinnaswamy, S. Confounding by Single Nucleotide Polymorphism rs117648444 (P70S) Affects the Association of Interferon Lambda Locus Variants with Response to Interferon-α-Ribavirin Therapy in Patients with Chronic Genotype 3 Hepatitis C Virus Infection. J. Interferon Cytokine Res. 2017, 37, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Higami, K.; Masaki, N.; Sugiyama, M.; Mukaide, M.; Saito, H.; Aoki, Y.; Sato, Y.; Imamura, M.; Murata, K.; et al. The rs8099917 Polymorphism, When Determined by a Suitable Genotyping Method, Is a Better Predictor for Response to Pegylated Alpha Interferon/Ribavirin Therapy in Japanese Patients than Other Single Nucleotide Polymorphisms Associated with Interleukin-28B. J. Clin. Microbiol. 2011, 49, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Riva, E.; Scagnolari, C.; Turriziani, O.; Antonelli, G. Hepatitis C virus and interferon type III (interferon-λ3/interleukin-28B and interferon-λ4): Genetic basis of susceptibility to infection and response to antiviral treatment. Clin. Microbiol. Infect. 2014, 20, 1237–1245. [Google Scholar] [CrossRef]
- Ibrahim, M.K.; AbdElrahman, M.; Din, N.G.B.; Tawfik, S.; Abd-Elsalam, S.; Omran, D.; Barakat, A.Z.; Farouk, S.; Elbatae, H.; El Awady, M.K. The impact of genetic variations in sofosbuvir metabolising enzymes and innate immunity mediators on treatment outcome in HCV-infected patients. Microb. Pathog. 2022, 162, 105311. [Google Scholar] [CrossRef]
- Loucks, C.M.; Lin, J.J.; Trueman, J.N.; Drögemöller, B.I.; Wright, G.E.B.; Chang, W.C.; Li, K.H.; Yoshida, E.M.; Ford, J.A.; Lee, S.S.; et al. Patient-specific genetic factors predict treatment failure in sofosbuvir-treated patients with chronic hepatitis C. Liver Int. 2022, 42, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Abdelnajid, D.; Elmowafy, A.Y.; Rostaing, L.; Elrakaiby, M.T. Prediction of response to sofosbuvir-based therapy using serum interleukin-12 and single nucleotide polymorphism of the interleukin 28B gene as predictive factors in HCV positive genotype-4 patients. Medicine 2023, 102, e34125. [Google Scholar] [CrossRef] [PubMed]
- Suppiah, V.; Moldovan, M.; Ahlenstiel, G.; Berg, T.; Weltman, M.; Abate, M.L.; Bassendine, M.; Spengler, U.; Dore, G.J.; Powell, E.; et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat. Genet. 2009, 41, 1100–1104. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nishida, N.; Sugiyama, M.; Kurosaki, M.; Matsuura, K.; Sakamoto, N.; Nakagawa, M.; Korenaga, M.; Hino, K.; Hige, S.; et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009, 41, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.H.; Bochud, P.-Y.; George, J. Host—Hepatitis C viral interactions: The role of genetics. J. Hepatol. 2016, 65, S22–S32. [Google Scholar] [CrossRef] [PubMed]
- Asthana, M.; Sahu, S.K.; Kumar, A.; Mohanty, S.; Chakrabarti, S.; Das, P.; Chattopadhya, N.R.; Chatterjee, K.; Singh, S.P.; Rajasubramaniam, S.; et al. Role of Interleukin 28B Polymorphisms in Response to Interferon Based Therapy for Hepatitis C Virus Clearance. Curr. Drug Metab. 2018, 19, 215–223. [Google Scholar] [CrossRef]
- Ansari, M.A.; Aranday-Cortes, E.; Ip, C.L.; da Silva Filipe, A.; Lau, S.H.; Bamford, C.; Bonsall, D.; Trebes, A.; Piazza, P.; Sreenu, V.; et al. Interferon lambda 4 impacts the genetic diversity of hepatitis C virus. eLife 2019, 8, e42463. [Google Scholar] [CrossRef]
- Ferreira, J.; Oliveira, M.; Bicho, M.; Serejo, F. Role of Inflammatory/Immune Response and Cytokine Polymorphisms in the Severity of Chronic Hepatitis C (CHC) before and after Direct Acting Antiviral (DAAs) Treatment. Int. J. Mol. Sci. 2023, 24, 1380. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Justice, A.C.; Marconi, V.C.; Aouizerat, B.E.; Xu, K. Co-occurrence of injection drug use and hepatitis C increases epigenetic age acceleration that contributes to all-cause mortality among people living with HIV. Epigenetics 2023, 18, 2212235. [Google Scholar] [CrossRef]
- Gao, Y.; Nepal, N.; Jin, S.Z. Toll-like receptors and hepatitis C virus infection. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 521–529. [Google Scholar] [CrossRef]
- Jiménez-Sousa, M.A.; Rallón, N.; Berenguer, J.; Pineda-Tenor, D.; López, J.C.; Soriano, V.; Guzmán-Fulgencio, M.; Cosín, J.; Retana, D.; García-Álvarez, M.; et al. TLR3 polymorphisms are associated with virologic response to hepatitis C virus (HCV) treatment in HIV/HCV coinfected patients. J. Clin. Virol. 2015, 66, 62–67. [Google Scholar] [CrossRef]
- Xu, Y.; Xue, W.; Gao, H.; Cui, J.; Zhao, L.; You, C. Association of toll-like receptors single nucleotide polymorphisms with HBV and HCV infection: Research status. PeerJ 2022, 10, e13335. [Google Scholar] [CrossRef]
- Talaat, R.M.; Elsayed, S.S.; Abdel-Hakem, N.E.; El-Shenawy, S.Z. Genetic Polymorphism in Toll-Like Receptor 3 and Interferon Regulatory Factor 3 in Hepatitis C Virus-Infected Patients: Correlation with Liver Cirrhosis. Viral Immunol. 2022, 35, 609–615. [Google Scholar] [CrossRef]
- Öksüz, Z.; Gragnani, L.; Lorini, S.; Temel, G.; Serin, M.S.; Zignego, A.L. Evaluation of Plasma miR-17-5p, miR-24-3p and miRNA-223-3p Profile of Hepatitis C Virus-Infected Patients after Treatment with Direct-Acting Antivirals. J. Pers. Med. 2023, 13, 1188. [Google Scholar] [CrossRef]
- Kulkarni, V.; Jayakumar, S.; Mohan, M.; Kulkarni, S. Aid or Antagonise: Nuclear Long Noncoding RNAs Regulate Host Responses and Outcomes of Viral Infections. Cells 2023, 12, 987. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, S.; Hashmi, A.H.; Khan, A.; Asad Raza Kazmi, S.M.; Manzoor, S. Emergence and Persistence of Resistance-Associated Substitutions in HCV GT3 Patients Failing Direct-Acting Antivirals. Front. Pharmacol. 2022, 13, 894460. [Google Scholar] [CrossRef] [PubMed]
- Sølund, C.; Pedersen, M.S.; Fahnøe, U.; Filskov, J.; Jenssen, H.; Weis, N.; Schønning, K.; Bukh, J. Pre-existing, treatment-specific resistance-associated substitutions in hepatitis C virus genotype 1 and 3 and viral RNA titers during treatment with direct-acting antivirals. APMIS 2023, 131, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Devan, P.; Tiong, K.L.A.; Neo, J.E.; Mohan, B.P.; Wijarnpreecha, K.; Tam, Y.C.S.; Coppola, N.; Preda, C.M.; Wong, Y.J. Treatment Outcomes of Sofosbuvir/Velpatasvir/Voxilaprevir in Direct-Acting Antiviral-Experienced Hepatitis C Virus Patients: A Systematic Review and Meta-Analysis. Viruses 2023, 15, 1489. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Li, L.; Hou, Z.; Zhang, Y.; Tang, Z.; Liu, S. Direct-acting Antiviral in the Treatment of Chronic Hepatitis C: Bonuses and Challenges. Int. J. Med. Sci. 2020, 17, 892–902. [Google Scholar] [CrossRef]
- Malandris, K.; Kalopitas, G.; Theocharidou, E.; Germanidis, G. The Role of RASs/RVs in the Current Management of HCV. Viruses 2021, 13, 2096. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.L. Global Elimination of Chronic Hepatitis. N. Engl. J. Med. 2019, 380, 2041–2050. [Google Scholar] [CrossRef]
- Garcia, A.; Fernandez, S.; Toro, F.; Sanctis, J. An Overview of Hepatitis C Vaccines. Recent Pat. Inflamm. Allergy Drug Discov. 2014, 8, 85–91. [Google Scholar] [CrossRef]
- Bailey, J.R.; Barnes, E.; Cox, A.L. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology 2019, 156, 418–430. [Google Scholar] [CrossRef]
- Duncan, J.D.; Urbanowicz, R.A.; Tarr, A.W.; Ball, J.K. Hepatitis C Virus Vaccine: Challenges and Prospects. Vaccines 2020, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Kardani, K.; Sadat, S.M.; Kardani, M.; Bolhassani, A. The next generation of HCV vaccines: A focus on novel adjuvant development. Expert Rev. Vaccines 2021, 20, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Keck, M.-L.; Wrensch, F.; Pierce, B.G.; Baumert, T.F.; Foung, S.K.H. Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine. Front. Immunol. 2018, 9, 1194. [Google Scholar] [CrossRef]
- Elbahrawy, A.; Atalla, H.; Alboraie, M.; Alwassief, A.; Madian, A.; El Fayoumie, M.; Tabll, A.A.; Aly, H.H. Recent Advances in Protective Vaccines against Hepatitis Viruses: A Narrative Review. Viruses 2023, 15, 214. [Google Scholar] [CrossRef]
- Adugna, A. Therapeutic strategies and promising vaccine for hepatitis C virus infection. Immun. Inflamm. Dis. 2023, 11, e977. [Google Scholar] [CrossRef]
- Fauvelle, C.; Colpitts, C.C.; Keck, Z.Y.; Pierce, B.G.; Foung, S.K.H.; Baumert, T.F. Hepatitis C virus vaccine candidates inducing protective neutralising antibodies. Expert Rev. Vaccines 2016, 15, 1535–1544. [Google Scholar] [CrossRef]
- Landi, A.; Law, J.; Hockman, D.; Logan, M.; Crawford, K.; Chen, C.; Kundu, J.; Ebensen, T.; Guzman, C.; Deschatelets, L.; et al. Superior immunogenicity of HCV envelope glycoproteins when adjuvanted with cyclic-di-AMP, a STING activator or archaeosomes. Vaccine 2017, 35, 6949–6956. [Google Scholar] [CrossRef]
- Lin, Y.; Kwon, T.; Polo, J.; Zhu, Y.-F.; Coates, S.; Crawford, K.; Dong, C.; Wininger, M.; Hall, J.; Selby, M.; et al. Induction of Broad CD4 + and CD8 + T-Cell Responses and Cross- Neutralizing Antibodies against Hepatitis C Virus by Vaccination with Th1-Adjuvanted Polypeptides Followed by Defective Alphaviral Particles Expressing Envelope Glycoproteins gpE1 and gpE2 and Nonstructural Proteins 3, 4, and 5. J. Virol. 2008, 82, 7492–7503. [Google Scholar]
- Gomez-Escobar, E.; Roingeard, P.; Beaumont, E. Current Hepatitis C Vaccine Candidates Based on the Induction of Neutralizing Antibodies. Viruses 2023, 15, 1151. [Google Scholar] [CrossRef]
- Alkrekshi, A.; Tamaskar, I. Safety of Immune Checkpoint Inhibitors in Patients with Cancer and Hepatitis C Virus Infection. Oncologist 2021, 26, e827–e830. [Google Scholar] [CrossRef]
- Patnaik, R.; Tsai, E. Hepatitis C Virus Treatment and Solid Organ Transplantation. Gastroenterol. Hepatol. 2022, 18, 85–94. [Google Scholar]
- Desombere, I.; Mesalam, A.A.; Urbanowicz, R.A.; Van Houtte, F.; Verhoye, L.; Keck, Z.Y.; Farhoudi, A.; Vercauteren, K.; Weening, K.E.; Baumert, T.F.; et al. A novel neutralising human monoclonal antibody broadly abrogates hepatitis C virus infection in vitro and in vivo. Antivir. Res. 2017, 148, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Thuru, X. Targeting of Tetraspanin CD81 with Monoclonal Antibodies and Small Molecules to Combat Cancers and Viral Diseases. Cancers 2023, 15, 2186. [Google Scholar] [CrossRef] [PubMed]
- Carriquí-Madroñal, B.; Lasswitz, L.; von Hahn, T.; Gerold, G. Genetic and pharmacological perturbation of hepatitis-C virus entry. Curr. Opin. Virol. 2023, 62, 101362. [Google Scholar] [CrossRef]
- Feld, J.J.; Cypel, M.; Kumar, D.; Dahari, H.; Ribeiro, R.V.P.; Marks, N.; Kamkar, N.; Bahinskaya, I.; Onofrio, F.Q.; Zahoor, M.A.; et al. Short-course, direct-acting antivirals and Ezetimibe to prevent HCV infection in recipients of organs from HCV-infected donors: A phase 3, single-centre, open-label study. Lancet. Gastroenterol. Hepatol 2020, 5, 649–657. [Google Scholar] [CrossRef]
- Gallard, C.; Lebsir, N.; Khursheed, H.; Reungoat, E.; Plissonnier, M.-L.; Bré, J.; Michelet, M.; Chouik, Y.; Zoulim, F.; Pécheur, E.-I.; et al. Heparanase-1 is upregulated by hepatitis C virus and favors its replication. J. Hepatol. 2022, 77, 29–41. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, F.; Zhou, H.; Quan, J.; Liu, C.; Wang, Y.; Xiao, F.; Liu, Q.; Liu, J.; Zhang, Y.; et al. Potential roles of heparanase in cancer therapy: Current trends and future direction. J. Cell. Physiol. 2023, 238, 896–917. [Google Scholar] [CrossRef]
- Saviano, A.; Habersetzer, F.; Lupberger, J.; Simo-Noumbissie, P.; Schuster, C.; Doffoël, M.; Schmidt-Mutter, C.; Baumert, T.F. Safety and Antiviral Activity of EGFR Inhibition by Erlotinib in Chronic Hepatitis C Patients: A Phase Ib Randomized Controlled Trial. Clin. Transl. Gastroenterol. 2022, 13, e00492. [Google Scholar] [CrossRef]
- Colpitts, C.C.; Tawar, R.G.; Mailly, L.; Thumann, C.; Heydmann, L.; Durand, S.C.; Xiao, F.; Robinet, E.; Pessaux, P.; Zeisel, M.B.; et al. Humanisation of a claudin-1-specific monoclonal antibody for clinical prevention and cure of HCV infection without escape. Gut 2018, 67, 736–745. [Google Scholar] [CrossRef]
- Pascut, D.; Hoang, M.; Nguyen, N.N.Q.; Pratama, M.Y.; Tiribelli, C. HCV Proteins Modulate the Host Cell miRNA Expression Contributing to Hepatitis C Pathogenesis and Hepatocellular Carcinoma Development. Cancers 2021, 13, 2485. [Google Scholar] [CrossRef]
- Mohamed, H.I.; Abdelrahim, E.M.; Elsayed, A.M.; Shaaban, S.M.; Eldahrouty, H.A. Relationship between hepatitis C virus infection and extrahepatic malignancies. Clinical and experimental hepatology. Clin. Exp. Hepatol. 2023, 9, 202–209. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina, C.; García, A.H.; Crespo, F.I.; Toro, F.I.; Mayora, S.J.; De Sanctis, J.B. A Synopsis of Hepatitis C Virus Treatments and Future Perspectives. Curr. Issues Mol. Biol. 2023, 45, 8255-8276. https://doi.org/10.3390/cimb45100521
Medina C, García AH, Crespo FI, Toro FI, Mayora SJ, De Sanctis JB. A Synopsis of Hepatitis C Virus Treatments and Future Perspectives. Current Issues in Molecular Biology. 2023; 45(10):8255-8276. https://doi.org/10.3390/cimb45100521
Chicago/Turabian StyleMedina, Christian, Alexis Hipólito García, Francis Isamarg Crespo, Félix Isidro Toro, Soriuska José Mayora, and Juan Bautista De Sanctis. 2023. "A Synopsis of Hepatitis C Virus Treatments and Future Perspectives" Current Issues in Molecular Biology 45, no. 10: 8255-8276. https://doi.org/10.3390/cimb45100521
APA StyleMedina, C., García, A. H., Crespo, F. I., Toro, F. I., Mayora, S. J., & De Sanctis, J. B. (2023). A Synopsis of Hepatitis C Virus Treatments and Future Perspectives. Current Issues in Molecular Biology, 45(10), 8255-8276. https://doi.org/10.3390/cimb45100521