Challenges in Defining a Reference Set of Differentially Expressed lncRNAs in Ulcerative Colitis by Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
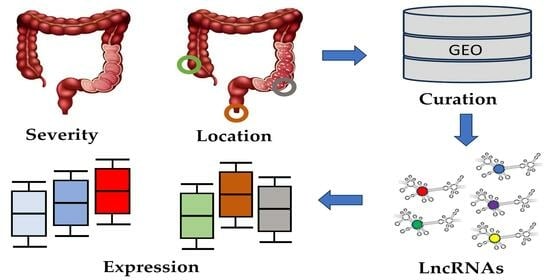
2.1. Selection of GEO Datasets and Samples
2.2. Dataset Curation
2.3. Data Processing
2.4. Expression of lncRNAs in Different Disease States and Tissue Locations
3. Results
3.1. The Number of Annotated LncRNA Gene Symbols Found in Each Dataset
3.2. Common LncRNA Gene Symbols Found in One to Nine Matrices
3.3. Differentially Expressed lncRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, P.; Chang, C.-P. Long Non-Coding RNA and Chromatin Remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and Tissue-Specific Transcription Patterns of Long Noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Mirza, A.H.; Berthelsen, C.H.; Seemann, S.E.; Pan, X.; Frederiksen, K.S.; Vilien, M.; Gorodkin, J.; Pociot, F. Transcriptomic Landscape of LncRNAs in Inflammatory Bowel Disease. Genome Med. 2015, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.; Carbonell-Sala, S.; De La Vega, F.M.; Faial, T.; Frankish, A.; Gingeras, T.; Guigo, R.; Harrow, J.L.; Hatzigeorgiou, A.G.; Johnson, R.; et al. The Status of the Human Gene Catalogue. arXiv 2023, arXiv:2303.13996v1. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, J. Are Human Translated Pseudogenes Functional? Mol. Biol. Evol. 2016, 33, 755–760. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, B.A.; Petrov, A.I.; Ribas, C.E.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Seemann, S.E.; Gorodkin, J.; Cannone, J.J.; et al. RNAcentral 2021: Secondary Structure Integration, Improved Sequence Search and New Member Databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar] [CrossRef]
- Yarani, R.; Mirza, A.H.; Kaur, S.; Pociot, F. The Emerging Role of Lncrnas in Inflammatory Bowel Disease. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative Colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Planell, N.; Lozano, J.J.; Mora-Buch, R.; Masamunt, M.C.; Jimeno, M.; Ordas, I.; Esteller, M.; Ricart, E.; Pique, J.M.; Panes, J.; et al. Transcriptional Analysis of the Intestinal Mucosa of Patients with Ulcerative Colitis in Remission Reveals Lasting Epithelial Cell Alterations. Gut 2013, 62, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Peyrin-Biroulet, L.; Bressenot, A.; Kampman, W. Histologic Remission: The Ultimate Therapeutic Goal in Ulcerative Colitis? Clin. Gastroenterol. Hepatol. 2014, 12, 929–934.e2. [Google Scholar] [CrossRef] [PubMed]
- Magro, F.; Gionchetti, P.; Eliakim, R.; Ardizzone, S.; Armuzzi, A.; Barreiro-de Acosta, M.; Burisch, J.; Gecse, K.B.; Hart, A.L.; Hindryckx, P.; et al. Third European Evidence-Based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, Diagnosis, Extra-Intestinal Manifestations, Pregnancy, Cancer Surveillance, Surgery, and Ileo-Anal Pouch Disorders. J. Crohn’s Colitis 2017, 11, 649–670. [Google Scholar] [CrossRef] [PubMed]
- Magro, F.; Langner, C.; Driessen, A.; Ensari, A.; Geboes, K.; Mantzaris, G.J.; Villanacci, V.; Becheanu, G.; Borralho Nunes, P.; Cathomas, G.; et al. European Consensus on the Histopathology of Inflammatory Bowel Disease. J. Crohns Colitis 2013, 7, 827–851. [Google Scholar] [CrossRef]
- Magro, F.; Lopes, J.; Borralho, P.; Lopes, S.; Coelho, R.; Cotter, J.; Castro, F.D.; Sousa, H.T.; Salgado, M.; Andrade, P.; et al. Comparison of Different Histological Indexes in the Assessment of UC Activity and Their Accuracy Regarding Endoscopic Outcomes and Faecal Calprotectin Levels. Gut 2018. [Google Scholar] [CrossRef] [PubMed]
- Olsen, T.; Goll, R.; Cui, G.; Husebekk, A.; Vonen, B.; Birketvedt, G.S.; Florholmen, J. Tissue Levels of Tumor Necrosis Factor-Alpha Correlates with Grade of Inflammation in Untreated Ulcerative Colitis. Scand. J. Gastroenterol. 2007, 42, 1312–1320. [Google Scholar] [CrossRef] [PubMed]
- Barreiro-de Acosta, M.; Vallejo, N.; De La Iglesia, D.; Uribarri, L.; Bastón, I.; Ferreiro-Iglesias, R.; Lorenzo, A.; Domínguez-Muñoz, J.E. Evaluation of the Risk of Relapse in Ulcerative Colitis According to the Degree of Mucosal Healing (Mayo 0 vs. 1): A Longitudinal Cohort Study. J. Crohn’s Colitis 2016, 10, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gulati, A.; Alipour, O.; Shao, L. Relapse From Deep Remission After Therapeutic De-Escalation in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. J. Crohn’s Colitis 2020, 14, 1413. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Cui, L.; Lin, C.; Wang, Y.; Liu, Z.; Miao, X. LncRNA CDKN2B-AS1 Relieved Inflammation of Ulcerative Colitis via Sponging MiR-16 and MiR-195. Int. Immunopharmacol. 2020, 88, 106970. [Google Scholar] [CrossRef] [PubMed]
- Sosnovski, K.E.; Braun, T.; Amir, A.; Moshel, D.; BenShoshan, M.; VanDussen, K.L.; Levhar, N.; Abbas-Egbariya, H.; Beider, K.; Ben-Yishay, R.; et al. GATA6-AS1 Regulates Intestinal Epithelial Mitochondrial Functions, and Its Reduced Expression Is Linked to Intestinal Inflammation and Less Favourable Disease Course in Ulcerative Colitis. J. Crohn’s Colitis 2023, 17, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Haidich, A.B. Meta-Analysis in Medical Research. Hippokratia 2010, 14, 29–37. [Google Scholar] [PubMed]
- Lee, Y.H. Strengths and Limitations of Meta-Analysis. Korean J. Med. 2019, 94, 391–395. [Google Scholar] [CrossRef]
- Haberman, Y.; Karns, R.; Dexheimer, P.J.; Schirmer, M.; Somekh, J.; Jurickova, I.; Braun, T.; Novak, E.; Bauman, L.; Collins, M.H.; et al. Ulcerative Colitis Mucosal Transcriptomes Reveal Mitochondriopathy and Personalized Mechanisms Underlying Disease Severity and Treatment Response. Nat. Commun. 2019, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Fenton, C.G.; Taman, H.; Florholmen, J.; Sørbye, S.W.; Paulssen, R.H. Transcriptional Signatures That Define Ulcerative Colitis in Remission. Inflamm. Bowel Dis. 2021, 27, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Pavlidis, P.; Tsakmaki, A.; Pantazi, E.; Li, K.; Cozzetto, D.; Digby- Bell, J.; Yang, F.; Lo, J.W.; Alberts, E.; Sa, A.C.C.; et al. Interleukin-22 Regulates Neutrophil Recruitment in Ulcerative Colitis and Is Associated with Resistance to Ustekinumab Therapy. Nat. Commun. 2022, 13, 5820. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Strauss, R.; Ouahed, J.; Chan, D.; Telesco, S.E.; Shouval, D.S.; Canavan, J.B.; Brodmerkel, C.; Snapper, S.B.; Friedman, J.R. Molecular Comparison of Adult and Pediatric Ulcerative Colitis Indicates Broad Similarity of Molecular Pathways in Disease Tissue. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Sandborn, W.J.; Feagan, B.G.; Marano, C.; Zhang, H.; Strauss, R.; Johanns, J.; Adedokun, O.J.; Guzzo, C.; Colombel, J.-F.; Reinisch, W.; et al. Subcutaneous Golimumab Induces Clinical Response and Remission in Patients with Moderate-to-Severe Ulcerative Colitis. Gastroenterology 2014, 146, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, X.; Li, G. Integrated Bioinformatics and Network Pharmacology to Identify the Therapeutic Target and Molecular Mechanisms of Huangqin Decoction on Ulcerative Colitis. Sci. Rep. 2022, 12, 159. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, J.T.; Nielsen, O.H.; Riis, L.B.; Pittet, V.; Mueller, C.; Rogler, G.; Olsen, J. Transcriptional Analysis of Left-Sided Colitis, Pancolitis, and Ulcerative Colitis-Associated Dysplasia. Inflamm. Bowel Dis. 2014, 20, 2340–2352. [Google Scholar] [CrossRef]
- Arijs, I.; De Hertogh, G.; Lemaire, K.; Quintens, R.; Van Lommel, L.; Van Steen, K.; Leemans, P.; Cleynen, I.; Van Assche, G.; Vermeire, S.; et al. Mucosal Gene Expression of Antimicrobial Peptides in Inflammatory Bowel Disease Before and After First Infliximab Treatment. PLoS ONE 2009, 4, e7984. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, W.; Peeters, P.M.; Staelens, D.; Schraenen, A.; Van der Goten, J.; Cleynen, I.; De Schepper, S.; Van Lommel, L.; Reynaert, N.L.; Schuit, F.; et al. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Fisher Exact Test—An Overview|ScienceDirect Topics. Available online: https://www.sciencedirect.com/topics/medicine-and-dentistry/fisher-exact-test (accessed on 30 November 2023).
- Jiang, C.; Li, Y.; Zhao, Z.; Lu, J.; Chen, H.; Ding, N.; Wang, G.; Xu, J.; Li, X. Identifying and Functionally Characterizing Tissue-Specific and Ubiquitously Expressed Human LncRNAs. Oncotarget 2016, 7, 7120–7133. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Sosnovski, K.E.; Amir, A.; BenShoshan, M.; VanDussen, K.L.; Karns, R.; Levhar, N.; Abbas-Egbariya, H.; Hadar, R.; Efroni, G.; et al. Mucosal Transcriptomics Highlight LncRNAs Implicated in Ulcerative Colitis, Crohn’s Disease, and Celiac Disease. JCI Insight 2023, 8, e170181. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.M.; Kim, E.; Ivanov, I.; Davidson, L.A.; Goldsby, J.S.; Hullar, M.A.; Randolph, T.W.; Kaz, A.M.; Levy, L.; Lampe, J.W.; et al. Comprehensive Site-Specific Whole Genome Profiling of Stromal and Epithelial Colonic Gene Signatures in Human Sigmoid Colon and Rectal Tissue. Physiol Genom. 2016, 48, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Van Beelen Granlund, A.; Flatberg, A.; Østvik, A.E.; Drozdov, I.; Gustafsson, B.I.; Kidd, M.; Beisvag, V.; Torp, S.H.; Waldum, H.L.; Martinsen, T.C.; et al. Whole Genome Gene Expression Meta-Analysis of Inflammatory Bowel Disease Colon Mucosa Demonstrates Lack of Major Differences between Crohn’s Disease and Ulcerative Colitis. PLoS ONE 2013, 8, e56818. [Google Scholar] [CrossRef]
- Linggi, B.; Jairath, V.; Zou, G.; Shackelton, L.M.; McGovern, D.P.B.; Salas, A.; Verstockt, B.; Silverberg, M.S.; Nayeri, S.; Feagan, B.G.; et al. Meta-Analysis of Gene Expression Disease Signatures in Colonic Biopsy Tissue from Patients with Ulcerative Colitis. Sci. Rep. 2021, 11, 18243. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Chen, L.-L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A Guide to Naming Human Non-coding RNA Genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef]
- Xu, J.; Shi, A.; Long, Z.; Xu, L.; Liao, G.; Deng, C.; Yan, M.; Xie, A.; Luo, T.; Huang, J.; et al. Capturing Functional Long Non-Coding RNAs through Integrating Large-Scale Causal Relations from Gene Perturbation Experiments. EBioMedicine 2018, 35, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Q.; Jiang, D.-M.; Hu, S.-S.; Zhao, L.; Wang, L.; Yang, M.-H.; Ai, M.-L.; Jiang, H.-J.; Han, Y.; Ding, Y.-Q.; et al. SATB2-AS1 Suppresses Colorectal Carcinoma Aggressiveness by Inhibiting SATB2-Dependent Snail Transcription and Epithelial–Mesenchymal Transition. Cancer Res. 2019, 79, 3542–3556. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Lagares, A.; Crujeiras, A.B.; Lopez-Serra, P.; Soler, M.; Setien, F.; Goyal, A.; Sandoval, J.; Hashimoto, Y.; Martinez-Cardús, A.; Gomez, A.; et al. Epigenetic Inactivation of the P53-Induced Long Noncoding RNA TP53 Target 1 in Human Cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E7535–E7544. [Google Scholar] [CrossRef] [PubMed]
- Pekow, J.; Meckel, K.; Dougherty, U.; Haider, H.I.; Deng, Z.; Hart, J.; Rubin, D.T.; Bissonnette, M. Increased Mucosal Expression of MiR-215 Precedes the Development of Neoplasia in Patients with Long-Standing Ulcerative Colitis. Oncotarget 2018, 9, 20709–20720. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, W.; Zhao, C.; Jiang, Q. LINC01224 Promotes Colorectal Cancer Progression by Sponging MiR-2467. Cancer Manag. Res. 2021, 13, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Huang, N.; Yin, Q.; Cheng, C.; Chen, D.; Gong, C.; Xiong, H.; Zhao, J.; Wang, J.; Li, X.; et al. LncRNA TP53TG1 Plays an Anti-Oncogenic Role in Cervical Cancer by Synthetically Regulating Transcriptome Profile in HeLa Cells. Front. Genet. 2022, 13, 981030. [Google Scholar] [CrossRef] [PubMed]
- Emam, O.; Wasfey, E.F.; Hamdy, N.M. Notch-Associated LncRNAs Profiling Circuiting Epigenetic Modification in Colorectal Cancer. Cancer Cell Int. 2022, 22, 316. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, X.-F.; Cheng, L.-N.; Li, X.-L. Long Non-Coding RNA CRNDE Promotes Cell Apoptosis by Suppressing MiR-495 in Inflammatory Bowel Disease. Exp. Cell Res. 2019, 382, 111484. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Duan, H.; Luo, H. Identification of Core Gene Expression Signature and Key Pathways in Colorectal Cancer. Front. Genet. 2020, 11, 505629. [Google Scholar] [CrossRef]
- Graham, L.D.; Pedersen, S.K.; Brown, G.S.; Ho, T.; Kassir, Z.; Moynihan, A.T.; Vizgoft, E.K.; Dunne, R.; Pimlott, L.; Young, G.P. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer 2011, 2, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Z.; Wu, Z.; Lin, W.; Yu, M. Downregulation of the Expression of the LncRNA MIAT Inhibits Melanoma Migration and Invasion through the PI3K/AKT Signaling Pathway. Cancer Biomark. 2019, 24, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.K.; Fenton, C.G.; Paulssen, R.H. Novel Long Non-Coding RNAs of Relevance for Ulcerative Colitis Pathogenesis. Non-Coding RNA Res. 2022, 7, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Arriaga-Canon, C.; Contreras-Espinosa, L.; Aguilar-Villanueva, S.; Bargalló-Rocha, E.; García-Gordillo, J.A.; Cabrera-Galeana, P.; Castro-Hernández, C.; Jiménez-Trejo, F.; Herrera, L.A. The Clinical Utility of LncRNAs and Their Application as Molecular Biomarkers in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 7426. [Google Scholar] [CrossRef] [PubMed]


| GEO Accession Number | PMID (Year) | UC Samples (N); (M/F) | Control Samples (N); (M/F) | Tissue | Platform | SSM |
|---|---|---|---|---|---|---|
| GSE109142 | 30604764 (2018) | 206 (112/94) | 20 (9/11) | rectal mucosal biopsy | Illumina HiSeq 2500 | NR |
| GSE128682 | 32322884 (2020) | 14 (9/5) | 16 (11/5) | sigmoid colon | NextSeq 550 | NR |
| GSE206285 | 36192482 (2022) | 550 (350/200) | 18 (9/9) | sigmoid colon | Affymetrix HT HG U133 + PM array | FFPE |
| GSE87466 | 29401083 (2018) | 87 (44/43) | 21 | 15–20 cm from anal verge | Affymetrix HT HG U133 + PM array | RNAlater |
| GSE92415 | 23735746 (2018) | 162 | 21 | colonic mucosal samples | Affymetrix HT HG U133 + PM array | NR |
| GSE107499 | NA (2018) | 59 (lesional) | 40 (non-lesional) | colon biopsy | Affymetrix Human Gene Expression Array | RNAlater |
| GSE47908 | 25358065 (2014) | 45 (20/25) | 15 (4/11) | descending colon | Affymetrix Human Genome U133 Plus 2.0 Arrays | RNA later/FFPE |
| GSE16879 | 19956723 (2009) | 24 (14/10) | 6 | colon | Affymetrix Human Genome U133 Plus 2.0 Arrays | NR |
| GSE59071 | 261692 (2015) | 97 | 11 | sigmoid or rectum | Affymetrix Human Gene 1.0 ST Array | snap-frozen |
| Datasets * | LncRNAs # |
|---|---|
| GSE107499 | 443 |
| GSE109142 | 2096 |
| GSE128682 | 4910 |
| GSE16879 | 2181 |
| GSE206285 | 2407 |
| GSE47908 | 2844 |
| GSE59071 | 778 |
| GSE87466 | 2843 |
| GSE92415 | 631 |
| LncRNA | GSE107499 | GSE10942 | GSE128682 | GSSE16879 | GSE206285 | GSE47908 | GSE59071 | GSE87466 | GSE92415 | sig_pct | nmat |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIR215 | N | S | S | N | N | N | S | N | N | 100 | 3 |
| DPP10-AS1 | N | S | S | Y | S | S | N | S | N | 83.3 | 6 |
| FAM30A | S | S | Y | S | S | S | Y | S | S | 77.8 | 9 |
| LINC02023 | N | N | N | Y | S | S | N | S | N | 75 | 4 |
| MIR155HG | N | S | S | N | N | Y | N | N | S | 75 | 4 |
| CDKN2B-AS1 | Y | S | S | Y | S | Y | N | S | S | 62.5 | 8 |
| VLDRL-AS1 | N | S | S | N | N | Y | Y | S | N | 60 | 5 |
| MIAT | N | S | Y | Y | S | Y | N | S | S | 57.1 | 7 |
| CRNDE | S | S | Y | Y | Y | N | N | N | S | 50 | 6 |
| FLI32255 | N | N | Y | Y | S | Y | N | S | S | 50 | 6 |
| GATA-AS1 | N | S | Y | Y | S | Y | N | S | N | 50 | 6 |
| LINC01215 | N | S | S | Y | Y | Y | N | S | N | 50 | 6 |
| LINC01224 | N | S | Y | Y | S | Y | N | S | N | 50 | 6 |
| MIR3936HG | N | N | Y | Y | S | Y | N | S | S | 50 | 6 |
| SATB2-AS1 | Y | S | Y | Y | S | Y | S | S | N | 50 | 8 |
| DIP2C-AS1 | Y | S | Y | Y | S | N | Y | N | S | 42.9 | 7 |
| FOXD2-AS1 | N | S | Y | Y | Y | Y | N | S | S | 42.9 | 7 |
| LINC03040 | S | S | S | Y | Y | Y | Y | Y | Y | 33.3 | 9 |
| TP53TG1 | Y | S | Y | Y | Y | Y | Y | S | S | 33.3 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenton, C.G.; Ray, M.K.; Paulssen, R.H. Challenges in Defining a Reference Set of Differentially Expressed lncRNAs in Ulcerative Colitis by Meta-Analysis. Curr. Issues Mol. Biol. 2024, 46, 3164-3174. https://doi.org/10.3390/cimb46040198
Fenton CG, Ray MK, Paulssen RH. Challenges in Defining a Reference Set of Differentially Expressed lncRNAs in Ulcerative Colitis by Meta-Analysis. Current Issues in Molecular Biology. 2024; 46(4):3164-3174. https://doi.org/10.3390/cimb46040198
Chicago/Turabian StyleFenton, Christopher G., Mithlesh Kumar Ray, and Ruth H. Paulssen. 2024. "Challenges in Defining a Reference Set of Differentially Expressed lncRNAs in Ulcerative Colitis by Meta-Analysis" Current Issues in Molecular Biology 46, no. 4: 3164-3174. https://doi.org/10.3390/cimb46040198
APA StyleFenton, C. G., Ray, M. K., & Paulssen, R. H. (2024). Challenges in Defining a Reference Set of Differentially Expressed lncRNAs in Ulcerative Colitis by Meta-Analysis. Current Issues in Molecular Biology, 46(4), 3164-3174. https://doi.org/10.3390/cimb46040198